


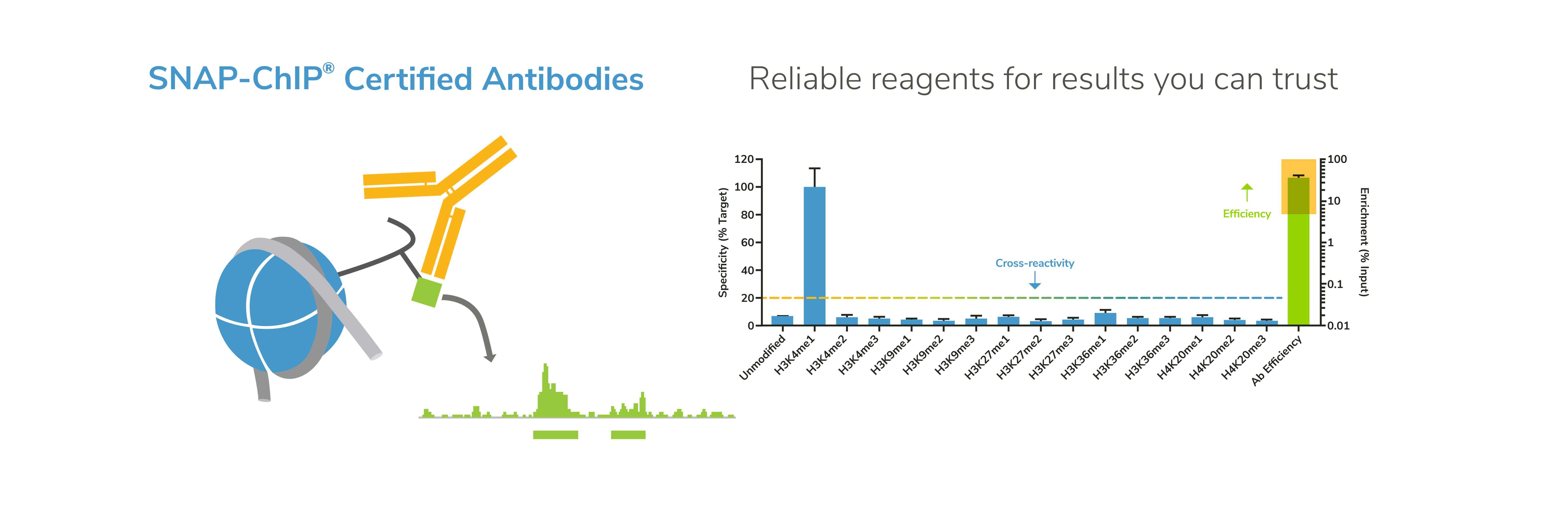
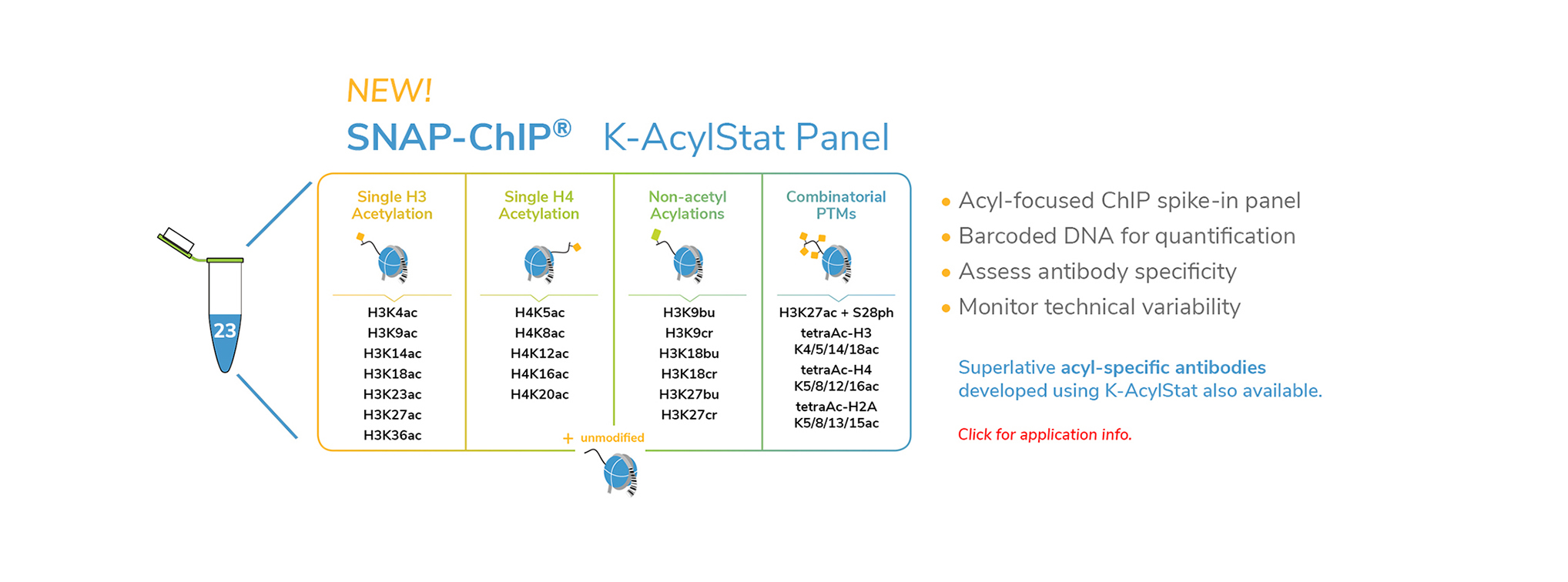
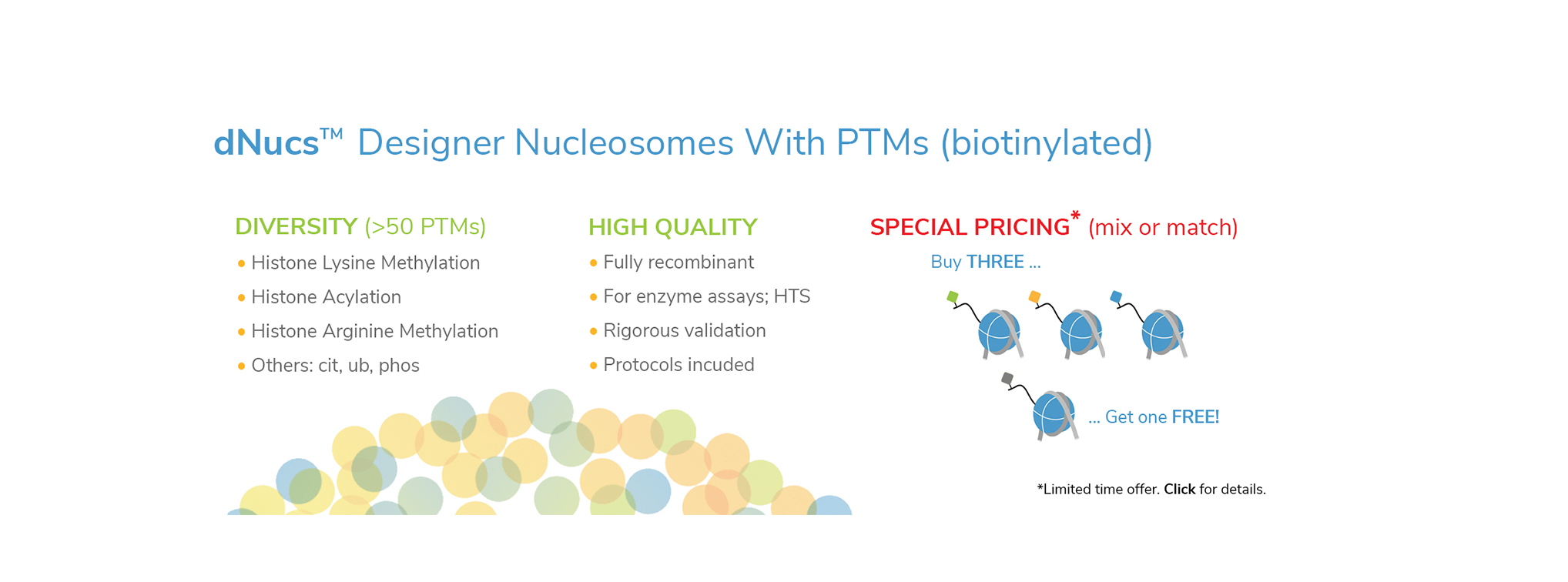
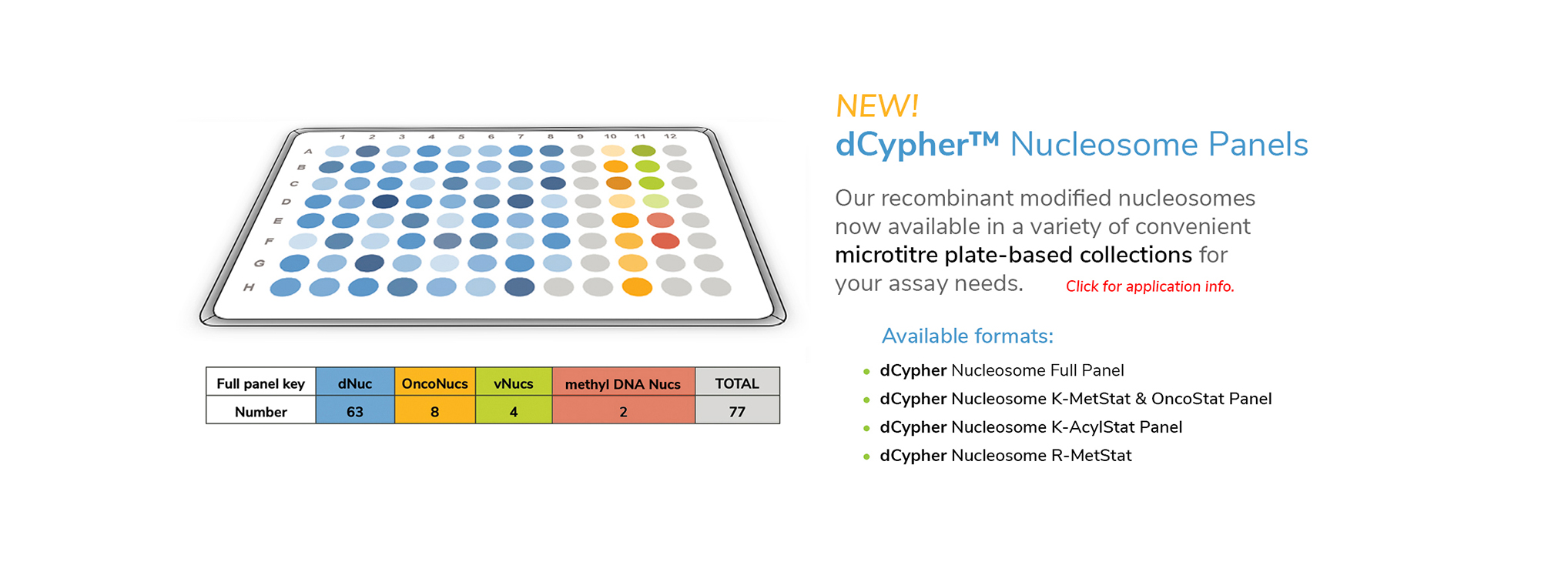

EpiCypher’s CUTANA™ CUT&RUN Services provide unique access to our genomic expertise, enabling diverse applications across biomedical research and drug development.
Learn MoreEpiDyne® enables direct detection of chromatin remodeling activity, and is ideal for HTS and inhibitor profiling. EpiDyne services accelerate assay development and generate robust results.
Learn MoredCypher™ assay services rapidly interrogate chromatin reader binding against a collection of modified histone peptides or nucleosomes, revealing novel mechanisms in chromatin biology.
Learn MoreNucleosomes are the preferred target of chromatin interacting proteins, and are optimal substrates for epigenetics research. To address this need, EpiCypher provides custom nucleosome development.
Learn More

