CUTANA™ CUT&RUN Library Prep Kit
Request a quote to save 10% when you buy our Library Prep and CUT&RUN Kits together (USA only)
The CUTANA™ CUT&RUN Library Prep Kit offers high fidelity library generation for Illumina® sequencing by harnessing the power of New England Biolabs® best-in-class NEBNext® reagents. The kit offers a streamlined protocol specifically optimized for high sensitivity CUT&RUN applications, including those with low cell inputs.
Included are all necessary reagents to perform end repair, adaptor ligation, combinatorial dual indexing for multiplexing up to 48 samples, and DNA cleanup with CUTANA DNA Purification Beads (EpiCypher 21-1407). If additional multiplexing is desired, Primer Sets 1 and 2 can be combined for sequencing of up to 96 samples in a single lane. Pairing this kit with the EpiCypher CUT&RUN Kit (EpiCypher 14-1048) affords users a cells-to-sequencing solution for chromatin mapping experiments with all the necessary controls and validated reagents to ensure confidence in obtaining high quality data. To place bulk orders, contact [email protected].
Trademarks and copyrights
New England Biolabs®, NEB®, and NEBNext® are registered trademarks of New England Biolabs, Inc.
This Product is covered by one or more patents, trademarks and/or copyrights owned or controlled by NEB. While NEB develops and validates its products for various applications, the use of this product may require the purchaser to obtain additional third-party intellectual property rights for certain applications.
This product is licensed for research and commercial use from Bio-Rad Laboratories, Inc., under U.S. Pat. Nos. 6,627,424, 7,541,170, 7,670,808, 7,666,645, and corresponding patents in other countries. No rights are granted for use of the product for Digital PCR or real-time PCR applications, with the exception of quantification in Next Generation Sequencing workflows.
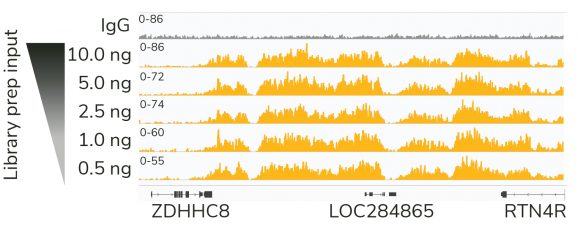
Figure 1: High quality sequencing libraries from as little as 0.5 ng of input
Gene browser shots generated using the Integrative Genomics Viewer (IGV, Broad Institute) show a representative 137 kb window for H3K27me3 antibody (ABclonal A16199) tested in CUT&RUN using 500k K562 cells. Decreasing inputs of CUT&RUN enriched DNA were used in the library prep protocol to simulate low abundance targets or low cell input experiments. Data are largely indistinguishable across inputs, demonstrating robust preparation of Illumina® NGS-ready libraries.
Figure 2: CUT&RUN DNA Fragment Size Distribution Analysis
CUT&RUN was performed using the CUTANA ChIC/CUT&RUN Kit (EpiCypher 14-1048) starting with 500k K562 cells. Five nanograms of CUT&RUN output DNA from reactions utilizing IgG (EpiCypher 13-0042), H3K4me3 (EpiCypher 13-0041), H3K27me3 (ABclonal A16199), and CTCF (EpiCypher 13-2014) antibodies were prepared for paired-end Illumina® sequencing with the CUTANA CUT&RUN Library Prep Kit. Library DNA was analyzed by Agilent TapeStation®, which confirmed that mononucleosomes were predominantly enriched in CUT&RUN (~300 bp peaks represent 150 bp nucleosomes + sequencing adapters). Peaks at ~380 bp correspond to the SNAP-CUTANA™ K-MetStat Panel of spike-in controls (EpiCypher 19-1002).
Figure 3: Representative gene browser tracks
CUT&RUN sequencing libraries described in Figure 2 were sequenced on an Illumina® NextSeq 2000 with a P3 cartridge (paired-end 2×50 cycle). Data were aligned to the hg19 genome using Bowtie2. Data were filtered to remove duplicates, multi-aligned reads, and ENCODE DAC Exclusion List regions. A representative 640 kb window at the SEPTIN5 gene is shown for three replicates (“Rep”) of IgG and H3K4me3 antibodies, as well as individual tracks for H3K27me3 and the transcription factor CTCF, demonstrating the robustness and reproducibility of the workflow with a variety of targets. Sequencing libraries prepared with the CUTANA CUT&RUN Library Prep kit produced the expected genomic distribution for each target. Sample sequencing depth was as follows (millions of reads): IgG Rep 1 (20.1), IgG Rep 2 (16.6), IgG Rep 3 (16.1), H3K4me3 Rep 1 (7.3), H3K4me3 Rep 2 (17.2), H3K4me3 Rep 3 (13.8), H3K27me3 (13.4), CTCF (16.2). Images were generated using the Integrative Genomics Viewer (IGV, Broad Institute).
Storage
OPEN KIT IMMEDIATELY and store components at room temperature and -20°C as indicated (see Kit Manual for full instructions). Stable for 6 months upon date of receipt.
Instructions for Use
See Kit Manual
| Item | Cat. No. |
|---|---|
| CUTANA™ CUT&RUN 8-strip 0.2 mL Tubes | 10-0009p |
| CUTANA™ DNA Purification Beads | 21-1407p |
| 0.1X TE Buffer | 21-1025p |
| End Prep Enzyme | 15-1019p |
| End Prep Buffer | 21-1012p |
| Adapter for Illumina® | 18-1000p |
| Ligation Mix | 15-1020p |
| Ligation Enhancer | 15-1021p |
| U-Excision Enzyme | 15-1023p |
| CUTANA™ Hot-Start 2X PCR Master Mix | 15-1022p |
| Multiplexing Primers | Primer Set 1 includes i701-i706 Primer Set 2 includes i707-i712 Both Sets include i501-i508 |
| Item | Cat. No. |
|---|---|
| CUTANA™ ChIC/CUT&RUN Kit | 14-1048 |
| CUT&RUN Antibodies | See the list |
| Magnetic Separation Rack, 0.2 mL Tubes | 10-0008 |