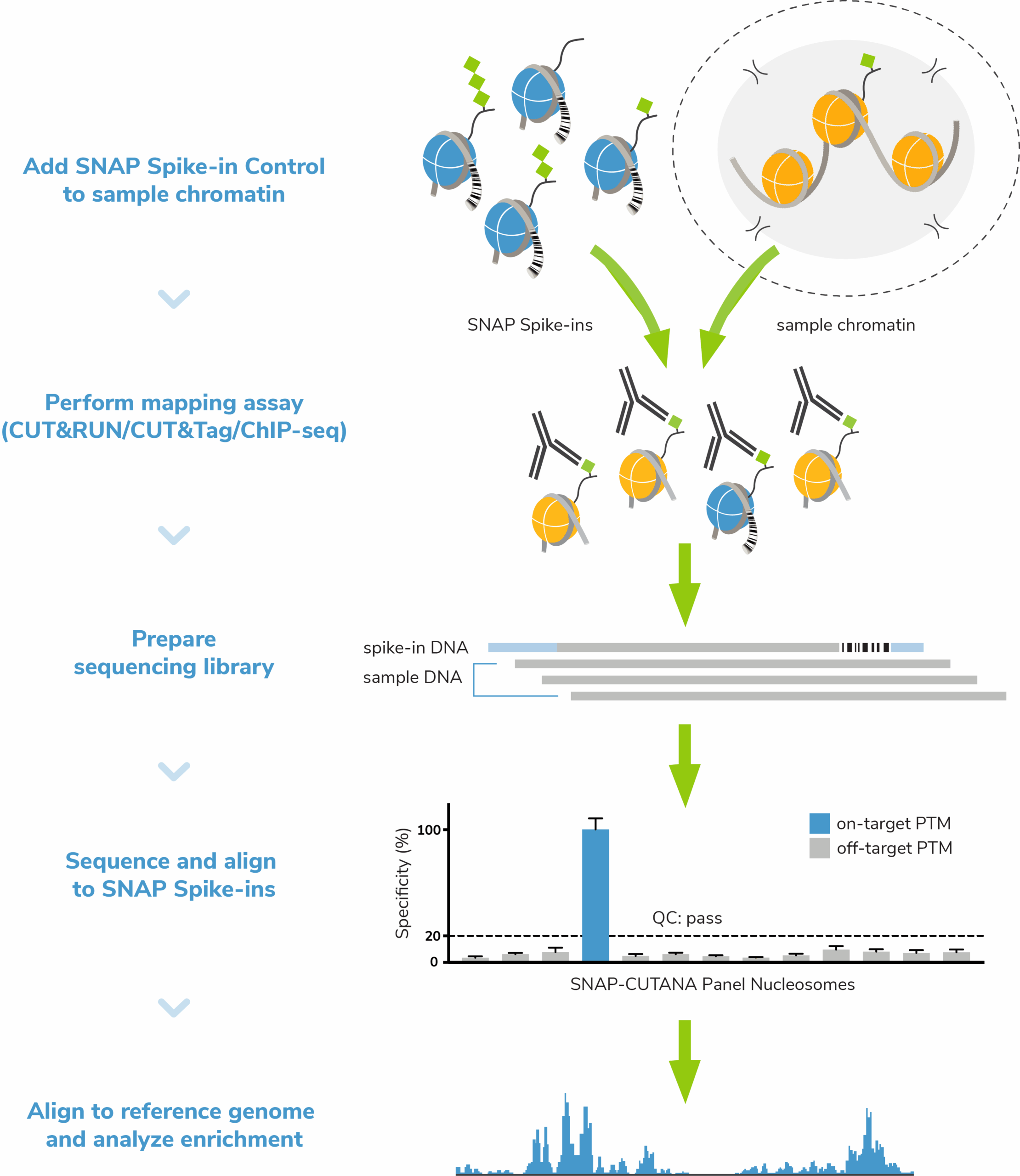
SNAP Spike-in Controls
All-in-one solution for quantitative chromatin profiling
Epigenomic mapping is a powerful and widely used approach for studying chromatin regulation. However, current controls are ill-defined and unable to provide accurate readouts of assay success, antibody specificity, or be used for quantitative normalization. The true quality of epigenomics data is often obscured, which misleads scientists and wastes valuable resources. EpiCypher has created SNAP Spike-in Controls to address these problems. With one easy step, you can control your assay and be confident in your data:
- Direct, quantitative readout of experimental success
- In-assay validation of antibody specificity
- Robust normalization for cross-sample comparisons
- Compatible with CUT&RUN, CUT&Tag and ChIP-seq assays
Have Questions?
We’re here to help. Click below and a member of our team will get back to you shortly!
Don't question your results — Save time and money with SNAP Spike-ins
What are SNAP Spike-in Controls?
SNAP Spike-in Controls are panels of defined recombinant human nucleosomes carrying widely studied histone post-translational modifications (PTMs) or common epitope tags, each wrapped with a barcoded DNA template that can be easily distinguished from sample chromatin by sequencing or qPCR.
SNAP Spike-ins are added to sample chromatin in one simple pipetting step at the beginning of your epigenomic mapping assay, subjecting them to all parts of the workflow and making them the ideal all-in-one control for chromatin profiling.
Spike-in panels are available for various chromatin mapping assays, including our CUTANA™ CUT&RUN and CUT&Tag assays, as well as ChIP-seq.
To learn more about our SNAP Spike-ins for CUT&RUN, check out our Technical Support Center.
The Advantages of SNAP Spike-ins
Quantitative controls for assay optimization & troubleshooting
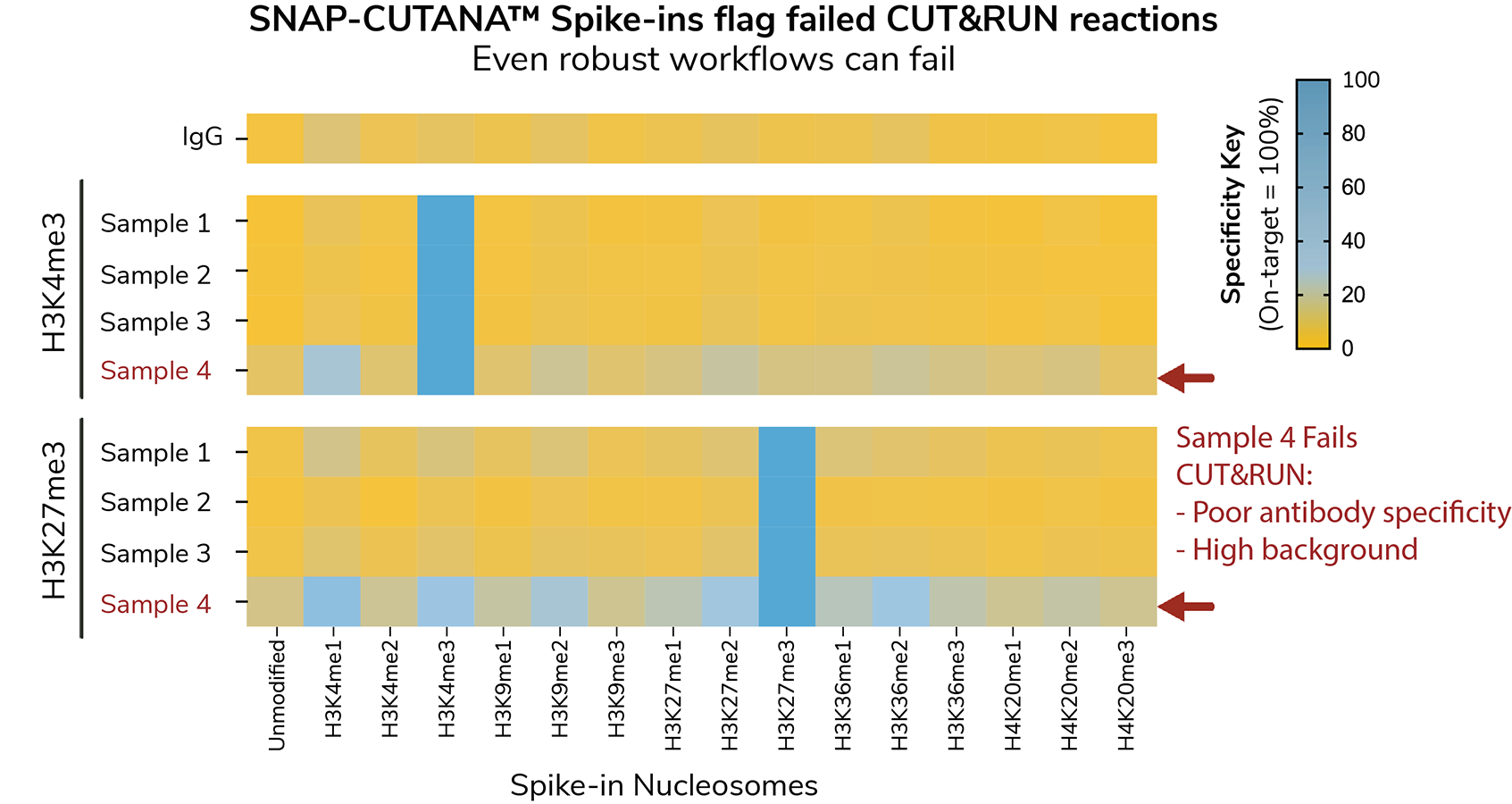
SNAP Spike-ins provide a direct readout of assay success. Comparing sample vs. spike-in data reveals valuable information about sample quality, antibody specificity, and workflow performance that is impossible with other controls. Use SNAP Spike-ins to:
- Flag failed reactions (see Figure)
- Guide troubleshooting: is the problem with cells, antibody, or workflow?
- Develop reliable assays for low cell inputs and clinical applications
- Continuously monitor assay success
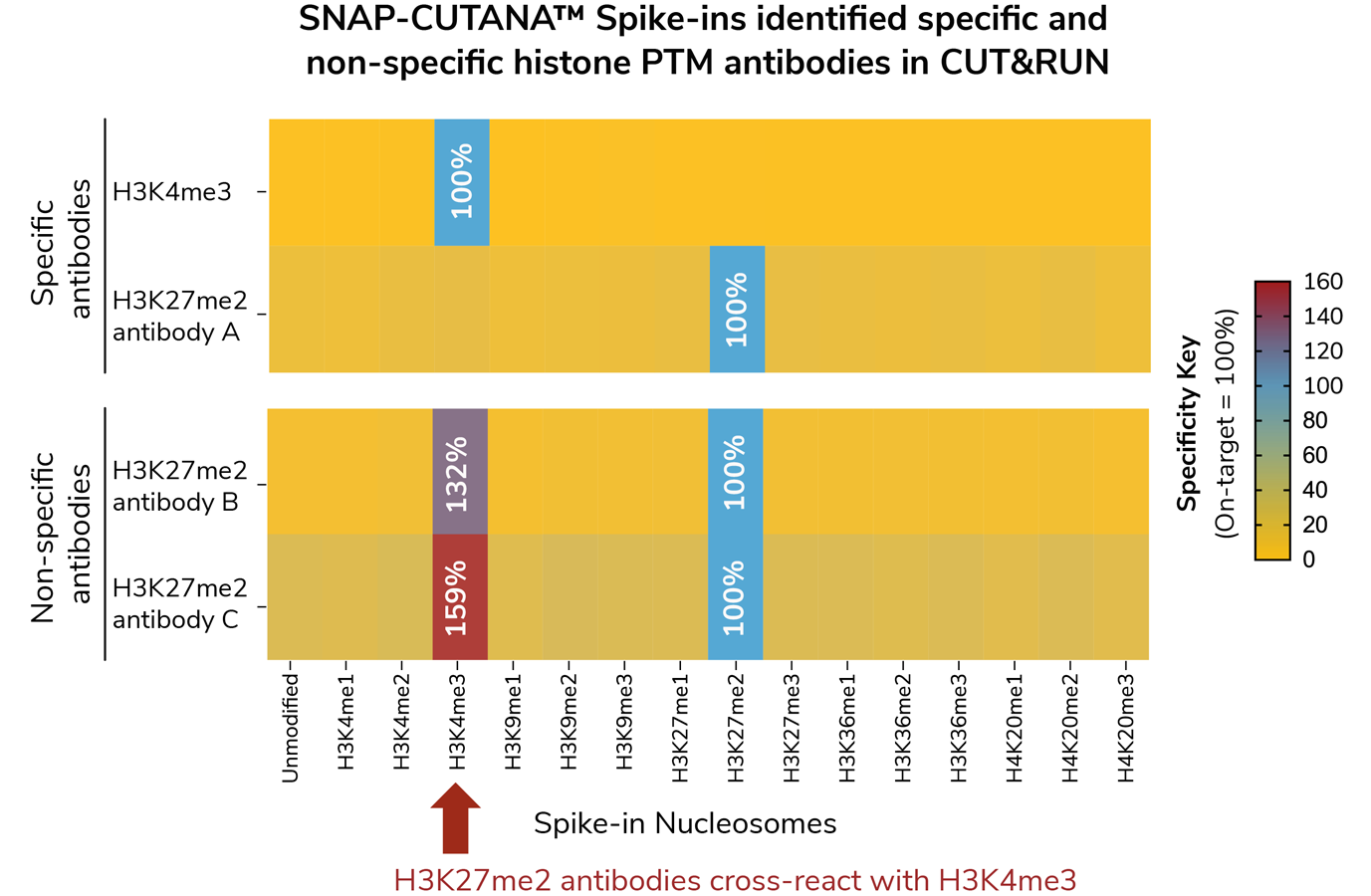
In-assay antibody validation against defined nucleosome controls
SNAP Spike-ins are the only control that allow you to directly test the specificity of your antibody where it matters most: in your assay.
- Direct readout of antibody specificity against highly pure physiological substrates
- Validate antibody for your unique cell type and conditions
- Avoid contaminating off-target signal for more accurate results (see Figure)
- Convenient panels of widely studied histone PTM targets (lysine methylation, lysine acylation, and more)
Robust sample normalization for reliable comparisons
Spike-in controls are essential to compare chromatin profiles across experiments, particularly for clinical and drug development applications. SNAP Spike-ins are a major advancement over existing exogenous chromatin spike-ins:
- Highly pure, lot-validated panels perform consistently across assays
- Compare data across samples, experiments and labs
- Standardize assay performance
- Quantify drug-induced changes in histone PTM enrichment that are obscured by standard methods (see Figure)
Interested in SNAP Spike-in Controls?

SNAP-CUTANA™ Spike-in Controls
Reliable nucleosome spike-in controls for quantitative CUT&RUN and CUT&Tag assays.
Explore Our Products
EpiCypher offers a wide range of products and services to aid you in your experimental needs. We have a growing catalog of spike-in controls (shown below), and certified antibodies. We also offer antibody validation as part of our CUTANA CUT&RUN Services – contact us to learn more. Related products include:
Featured Publications
- Shah et al. Examining the roles of H3K4 methylation states with systematically characterized antibodies. Mol Cell. 72, 162 – 177 (2018). (PMID: 30244833)
In this study, SNAP-ChIP Spike-in technology revealed that many widely cited, “ChIP-grade” antibodies exhibit poor binding specificity and pull-down efficiency, and demonstrated that histone peptide arrays do not accurately predict antibody performance. - Lam et al. Cell-type-specific genomics reveals histone modification dynamics in mammalian meiosis. Nat. Commun. 10, 3821 (2019). (PMID: 31444359)
SNAP-ChIP K-MetStat spike-ins were used to normalize ChIP-seq data and uncover epigenetic changes associated with meiosis. In choosing an antibody for their study, Lam and colleagues recapitulated findings from Shah et al., in which widely cited H3K4me3 antibody produced biologically distinct findings compared to a highly specific EpiCypher SNAP-ChIP Certified H3K4me3 antibody. - Grzybowski et al. Native internally calibrated chromatin immunoprecipitation for quantitative studies of histone post – translational modifications. Nat. Protoc. 14, 3275-3302 (2019). (PMID: 31723301) This study includes detailed methods for the use of barcoded recombinant nucleosomes as spike-in controls for ChIP normalization, and are directly applicable to the use of SNAP-ChIP spike-ins.
- Tay et al. Hdac3 is an epigenetic inhibitor of the cytotoxicity program in CD8 T cells. J. Exp. Med. 217, e20191453 (2020). (PMID: 32374402)
Reads from the DNA barcodes on SNAP-ChIP K-AcylStat spike-ins were used to normalize H3K27ac ChIP-seq data and elucidate H3K27ac changes after HDAC3 knockout in CD8+ T cells. Shirane et al. NSD1-deposited H3K36me2 directs de novo methylation in the mouse male germline and counteracts Polycomb-associated silencing. Nature Genetics 52, 1088-1098 (2020). (PMID: 32929285) Here, the Lorincz lab used SNAP-ChIP Certified Antibodies and Spike-in Controls to characterize the histone lysine methylation landscape in mouse male germline cells. These studies revealed a novel role for the H3K36 methyltransferase NSD1 in driving sexually dimorphic DNA methylation patterns in the germline.
Related Blog Posts

- Nathaniel Wesley
What is CUT&RUN? CUT&RUN, or Cleavage Under Targets and Release Using Nuclease, is a groundbreaking approach for ultra-sensitive genomic mapping...

- Nathaniel Wesley
Even if you are new to epigenetics, you have probably heard of ChIP-seq, CUT&RUN, and CUT&Tag – but how do...