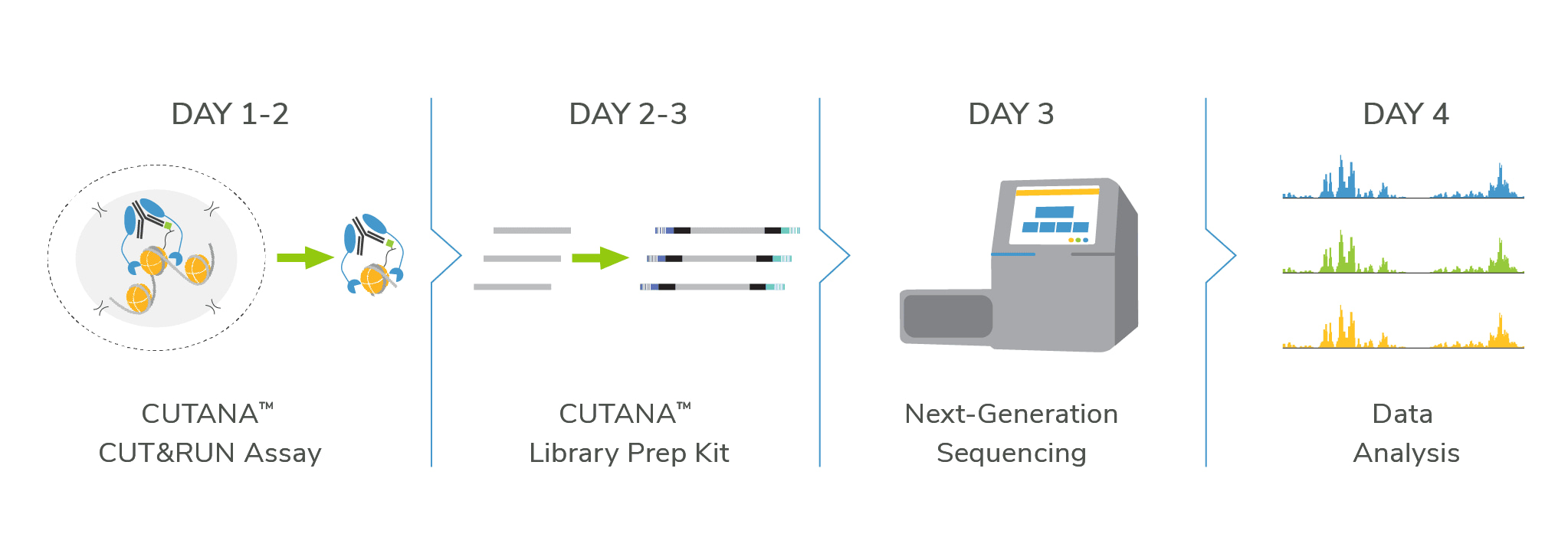
Gene expression is controlled by chromatin, a complex molecular landscape whose structure and function are determined by histone post-translational modifications (PTMs), transcription factors, and other chromatin-associated proteins. Mapping the genome-wide location of histone PTMs and chromatin-associated proteins is essential for exploring the epigenetic mechanisms linking chromatin regulation to gene expression and understanding how these pathways impact development and disease.
CUT&RUN is the leading epigenomic mapping assay, featuring unmatched throughput, versatility, and sensitivity. Whether you are new or experienced in epigenomics, starting a new technique can be intimidating. That’s why we put together this CUT&RUN overview, where you can learn everything you need to know about getting started with CUT&RUN.
What is CUT&RUN?
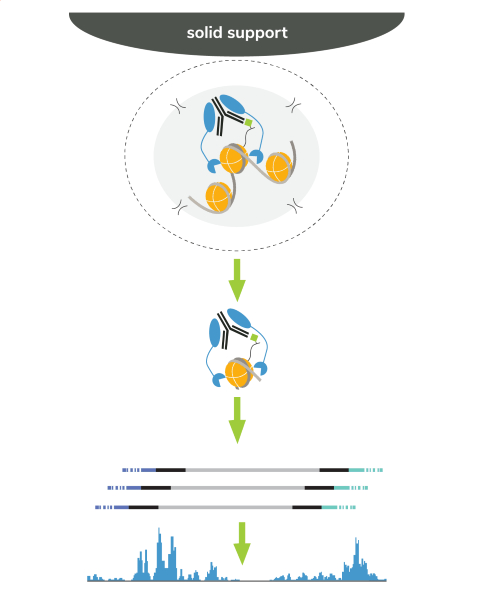
Cleavage Under Targets & Release Using Nuclease (CUT&RUN) is an epigenomic mapping assay that uses a Protein A/G Micrococcal Nuclease fusion protein (pAG-MNase) to selectively cleave antibody-bound chromatin. The entire assay takes place in intact cells or nuclei bound to magnetic beads, a solid support that streamlines sample processing. Following MNase activation, clipped chromatin fragments diffuse into solution while cells are easily removed using magnetic beads – eliminating a major source of background signal. Target-enriched DNA is then purified from the solution and used to prepare sequencing libraries.
CUT&RUN allows high-resolution epigenomic mapping for histone PTMs and chromatin-associated proteins using a small number of cells and only 3-8 million sequencing reads per reaction. The streamlined workflow and cost savings make CUT&RUN amenable to greater experimental throughput, allowing deeper and more rapid investigations to uncover epigenetic biology
How does CUT&RUN work?
CUT&RUN requires a few basic steps:
1. Sample preparation. Cells (or nuclei) are bound to Concanavalin A (ConA) magnetic beads and permeabilized with Digitonin to facilitate antibody and pAG-MNase binding.
2. Antibody binding. A target-specific antibody is added, which binds the chromatin feature of interest. Cells are washed several times following incubation to reduce nonspecific antibody binding.
3. Targeted cleavage using pAG-MNase. pAG-MNase is added to the reaction. Protein A/G binds immunoglobulin proteins, tethering MNase to antibody-bound chromatin loci. Calcium is added to activate MNase, which cleaves nearby genomic DNA.
4. DNA purification and sequencing. Digested chromatin fragments (antibody-bound complex; see figure) are released into solution and bead-bound cells are removed using a magnet. Target-enriched DNA is purified and prepared for sequencing
For further introduction to CUT&RUN, watch the video below or see this blog.
What is the difference between CUT&RUN and ChIP-seq?
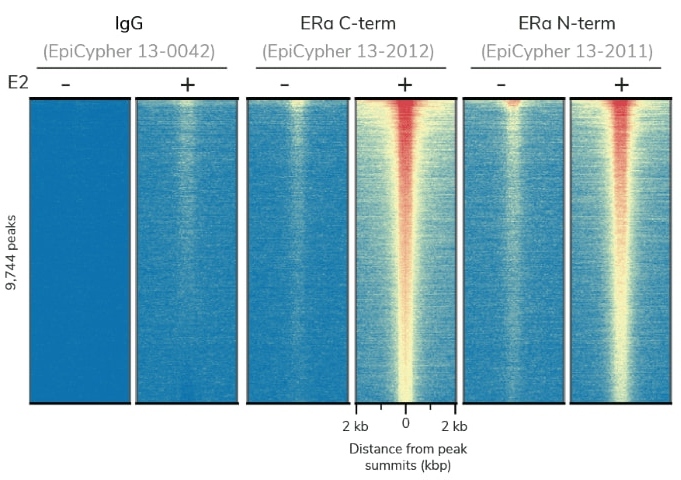
The main difference between ChIP-seq and CUT&RUN is how chromatin fragments are enriched. ChIP-seq (Chromatin ImmunoPrecipitation and sequencing) uses antibodies to “pull-down” target-associated DNA from a large pool of fragmented chromatin, resulting in high background and variable yields. In contrast, CUT&RUN uses pAG-MNase to selectively cleave antibody-bound chromatin in intact cells. Target-bound DNA diffuses into solution, leaving background behind – resulting in an assay with improved signal-to-noise and sensitivity.
What are the challenges of ChIP-seq?
ChIP-seq is a time-intensive and expensive technique with multiple problematic steps:
- Crosslinking: required to stabilize targets on chromatin, but causes artifacts
- Cellular lysis: makes samples viscous, hard to work with, and increases background
- Chromatin fragmentation: laborious, requires optimization, introduces variability
- Immunoprecipitation: requires stringent washes, diminishing signal-to-noise
To overcome these challenges, ChIP-seq requires high numbers of starting cells, deep sequencing, and complicated optimization steps. These requirements and the associated costs have precluded chromatin mapping for many applications, such as clinical and translational research, and slowed the development of effective epigenetics-targeted therapeutics.
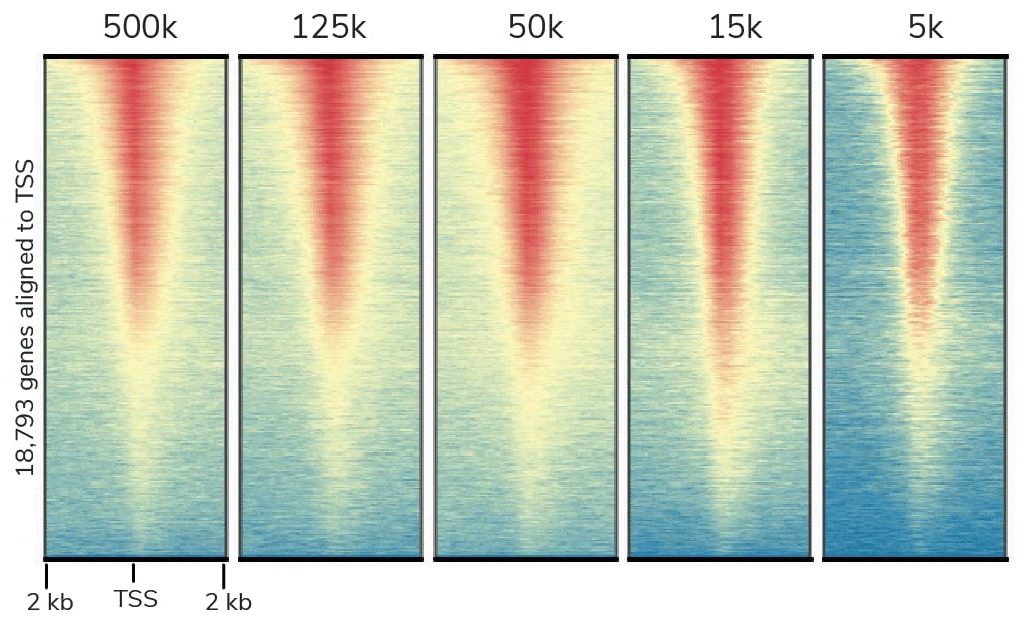
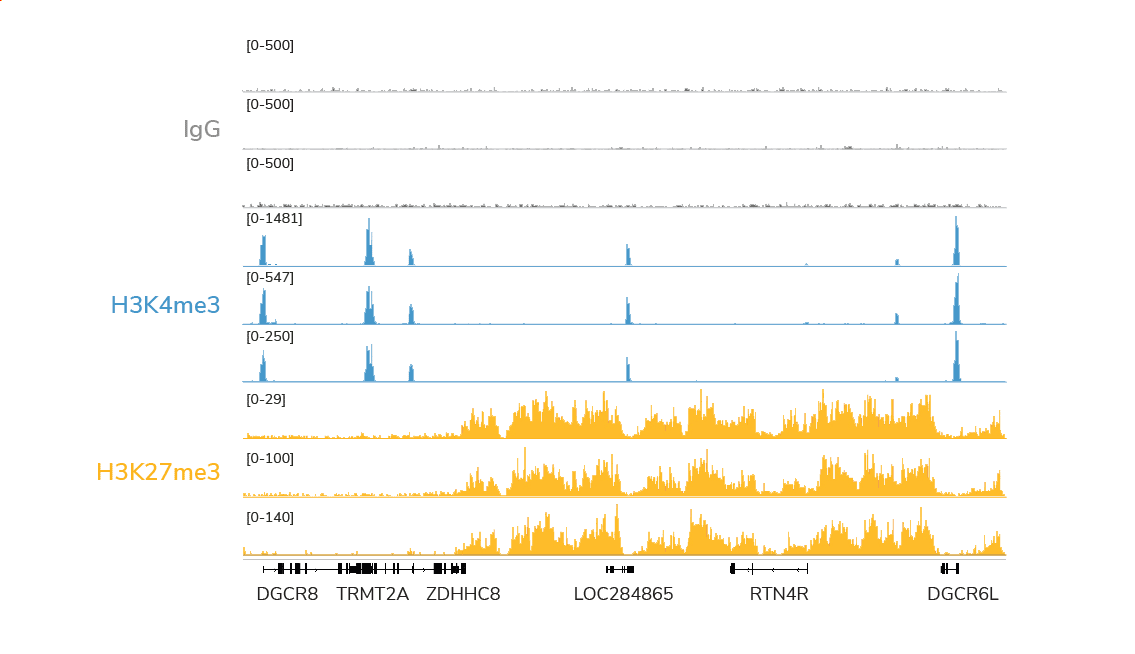
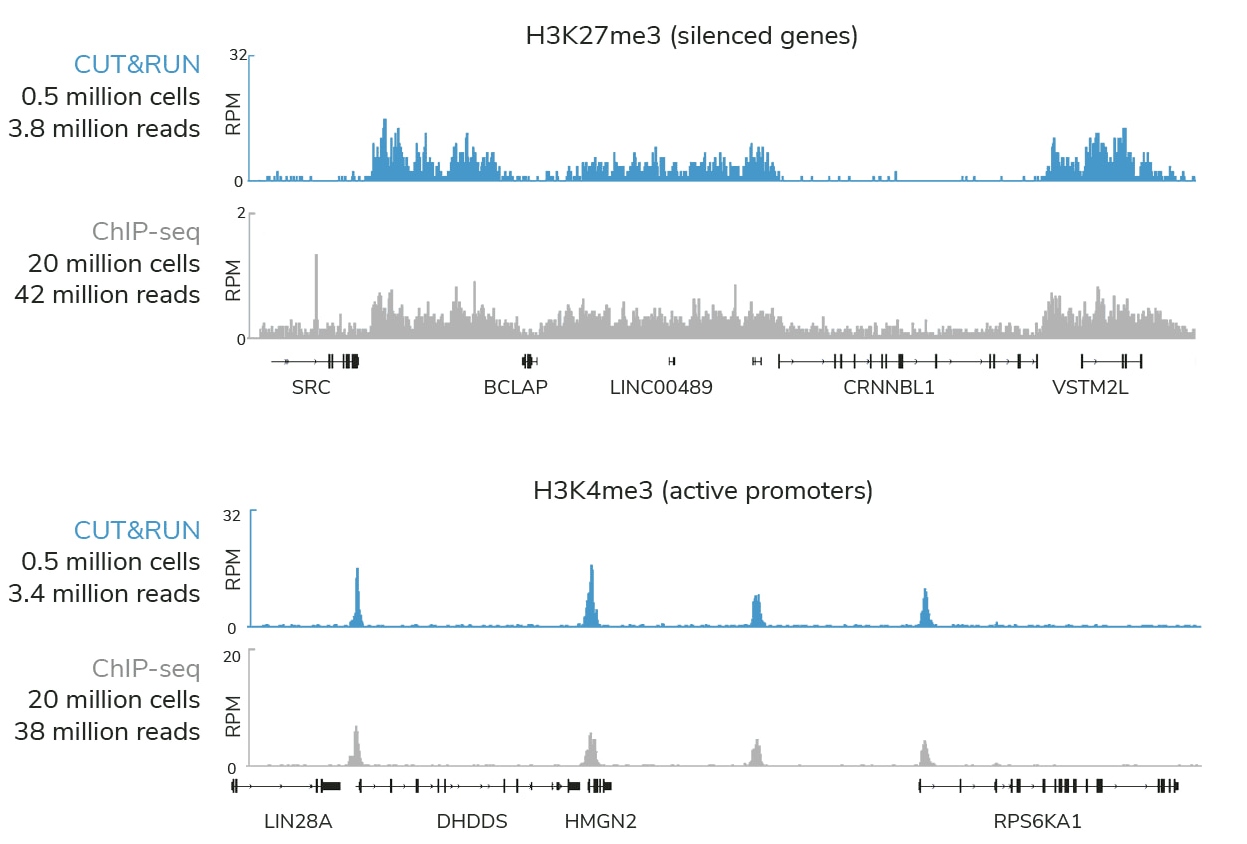 CUTANA™ CUT&RUN profiling using 0.5 million K562 cells and < 4 million sequencing reads
generates superior genomic maps for markers of silenced genes and active promoters
CUTANA™ CUT&RUN profiling using 0.5 million K562 cells and < 4 million sequencing reads
generates superior genomic maps for markers of silenced genes and active promoters
All of these challenges are eliminated in CUT&RUN.
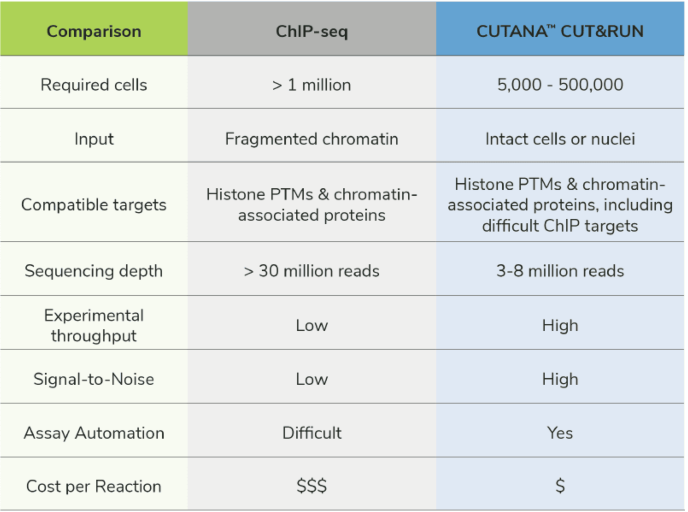
Compared to ChIP-seq, CUT&RUN delivers superior genomic mapping data using up to 40-fold fewer cells and >10-fold reduced sequencing depth (below). The user-friendly CUT&RUN workflow can go from cells to sequencing libraries in just 3 days and requires minimal optimization steps, all at a fraction of the costs of ChIP-seq.
Is CUT&RUN better than ChIP-seq?
CUT&RUN is superior to ChIP-seq in terms of cell input, sequencing depth, throughput, and cost. CUT&RUN assays are also more user-friendly, generate more reliable data, and work with a a diverse array of sample types, processing methods, and targets.
A full comparison of CUT&RUN technology and ChIP-seq can be found in the camparison table.
Advantages of CUT&RUN
CUT&RUN excels where ChIP-seq fails – providing a fast, reliable assay compatible with multiple sample and target types. Learn more about the capabilities of CUT&RUN below.
Low-input chromatin mapping
CUT&RUN enables epigenomic analysis of low-abundance targets and rare sample types, opening new avenues of research. With CUT&RUN:
- Produce high on-target, low background epigenomic maps
- Profile historically inaccessible cell types (biopsies, FACS-isolated cells)
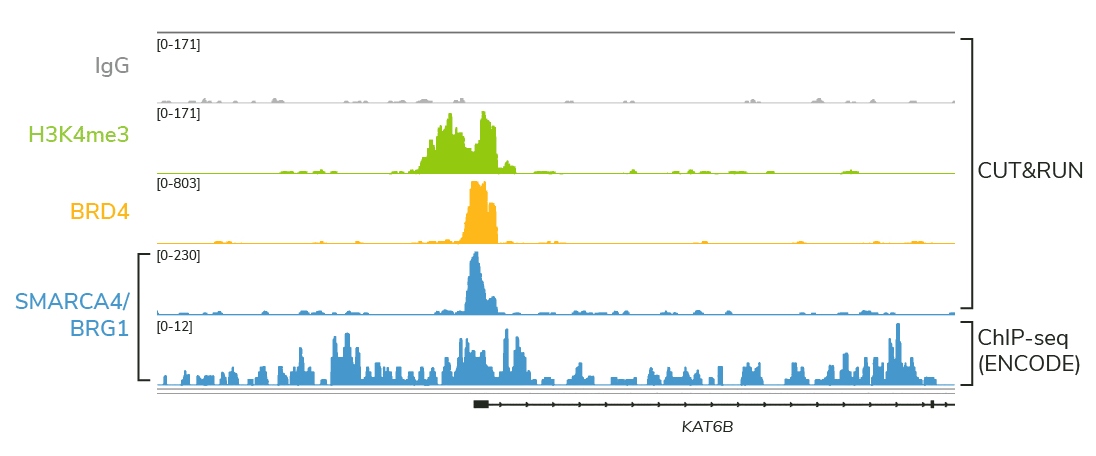
Map challenging targets with ease
Dysregulation of histone PTMs and chromatin-bound proteins is strongly linked to disease, making them key biomedical targets. Use CUT&RUN to study:
- Nucleosome remodelers – inaccessible by ChIP!
- Transcription factors, chromatin readers, and histone PTMs
Reveal dynamic chromatin changes
Chromatin-interacting proteins are emerging drug targets, but validating their therapeutic potential remains challenging. Ultra-sensitive CUT&RUN assays can:
- Detect epigenomic changes in response to drugs or stimuli
- Uncover novel drug targets or explore therapeutic mechanism of action
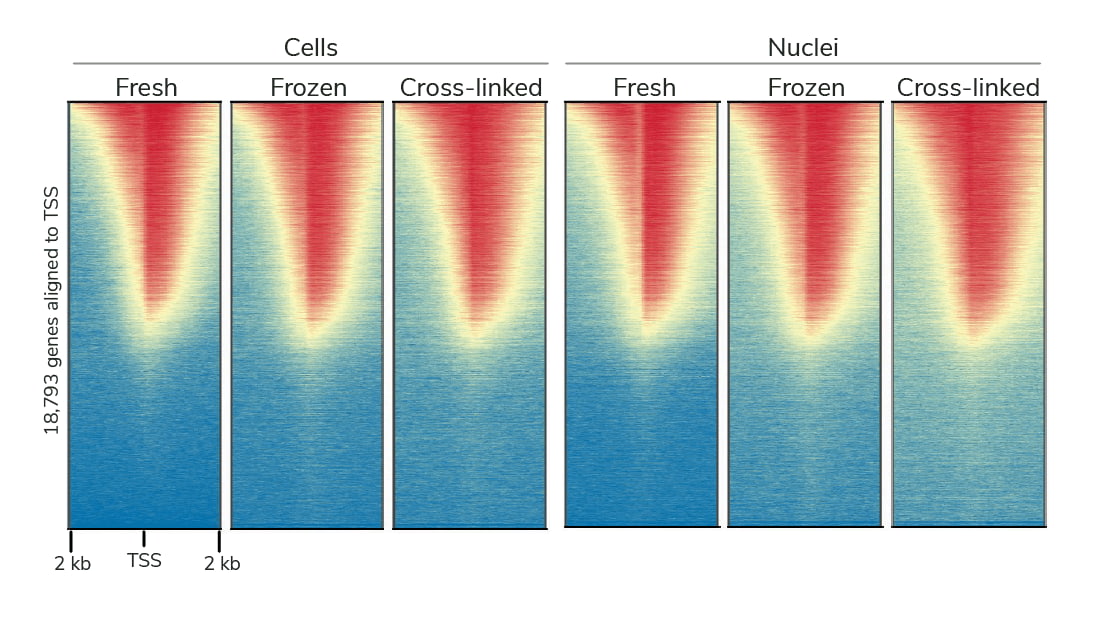
Broad sample type compatibility
CUT&RUN has been used in various research areas (see Key Publications and Example Applications). Specifically, CUT&RUN is:
- Compatible with fresh, frozen, or fixed cell lines, primary cells, and tissues
- Flexible to meet the needs of your project – including those using archived material
CUT&RUN Frequently Asked Questions
Interested in CUT&RUN but have questions about how it will integrate into your studies? Read on.
I am new to genomics, is CUT&RUN for me?
Yes! CUT&RUN is a robust, reliable, user-friendly genomic mapping assay. Our CUTANA CUT&RUN and Library Prep kits provide everything you need to integrate CUT&RUN in your research.
Does CUT&RUN require cross-linking?
No, unlike ChIP-seq, CUT&RUN does not require cross-linking. The preferred input for CUT&RUN is native (unfixed) samples. In fact, heavy cross-linking as with ChIP-seq severely reduces CUT&RUN yields and is not recommended for CUT&RUN under any circumstances.
However, EpiCypher has found that light cross-linking can improve experimental success for some labile histone PTMs, such as acetylation, or other chromatin-associated proteins. Full experimental guidance on how to test cross-linking for a specific target can be found on our protocols page.
What sample types is CUT&RUN compatible with?
CUTANA CUT&RUN is compatible with a vast array of cell types:
Species: Human, mouse, Drosophila, zebrafish, and more
Cell processing: Cultured cells (adherent and suspension), FACS-isolated cells, frozen samples
Cell types: Cell lines, differentiated pluripotent stem cells, primary cells (e.g. immune cells*), whole tissues (e.g. brain)
*Note on immune cells: ConA is a lectin and may activate some immune cell types. We recommend using nuclei if performing CUT&RUN with immune cells.
For examples, see Key Publications and Example Applications below. Guidance on sample prep, including protocol modifications, can be found in the Tech Support Center.
Is CUT&RUN compatible with whole tissues?
Yes, CUT&RUN is compatible with tissues. Tissues must first be processed into a single-cell suspension by mechanical or enzymatic means before being bound to ConA beads. The Tech Support Center provides more guidance on these methods.
How many cells do you need for CUT&RUN?
EpiCypher’s CUTANA CUT&RUN assays are validated for 500,000 to 5,000 cells*. Note that some targets that provide robust CUT&RUN data at 500,000 may require additional optimization or even fail at low inputs. Full considerations for low-input CUT&RUN experiments can be found on the Tech Support Center.
*Using fewer than 5,000 cells comes with many caveats, including low yields, increased adapter dimer contamination in libraries, and increased sequencing depth requirements.
Will CUT&RUN work for my ChIP target?
Yes, most likely. To date, we’ve confirmed that all major target classes – including histone PTMs, transcription factors, chromatin readers, writers, and modifying enzymes (erasers, remodelers) – generate robust data in CUT&RUN, all you need is a good antibody!
Can I use my ChIP-validated antibody for CUT&RUN?
No, we do not recommend using ChIP-validated antibodies for CUT&RUN as they may perform poorly in CUT&RUN due to the differences between these two assays. In general, EpiCypher has found that antibody validation in ChIP, immunoblot, ELISA or any other method does not predict performance in CUT&RUN. The only way to know if your antibody works for CUT&RUN is to test it in CUT&RUN.
EpiCypher offers a suite of CUT&RUN-validated antibodies for histone PTMs and chromatin associated proteins. If we do not offer a CUT&RUN-validated antibody for your target, see the advice in this blog post or contact us.
Should I use CUT&RUN or CUT&Tag?
EpiCypher has extensive experience in developing CUT&RUN, CUT&Tag, and ChIP-seq assays. CUT&RUN is our all-purpose chromatin mapping assay as it balances cell input and sequencing requirements with versatile target compatibility, including chromatin-associated proteins. For a deeper discussion of what to consider when choosing between CUT&RUN and CUT&Tag, see this blog post.
Why choose CUTANA CUT&RUN?
EpiCypher scientists extensively optimized each step of the CUT&RUN workflow and hand selected the best-performing companion products. The result is our collection of user-friendly and cost-effective CUTANA CUT&RUN and Library Prep kits, antibodies, controls, and supporting reagents, making this ground-breaking technology accessible to researchers worldwide.
All-in-one Kits for CUT&RUN and Library Prep
Our CUT&RUN and Library Prep kits deliver a streamlined solution for CUT&RUN experiments.
- The CUTANA CUT&RUN kit includes everything you need to go from cells to DNA, including an extensive manual and key experimental controls
- CUTANA CUT&RUN Library Prep kit harness the power of NEBNext® reagents and are specifically optimized for CUT&RUN experiments
Robust CUT&RUN protocol
EpiCypher’s user-friendly CUT&RUN protocol is optimized for:
- Diverse targets and samples
- Low cell numbers
- Streamlined profiling
- High sensitivity at low sequencing depths
In addition, we provide technical guidance to help you achieve experimental success.
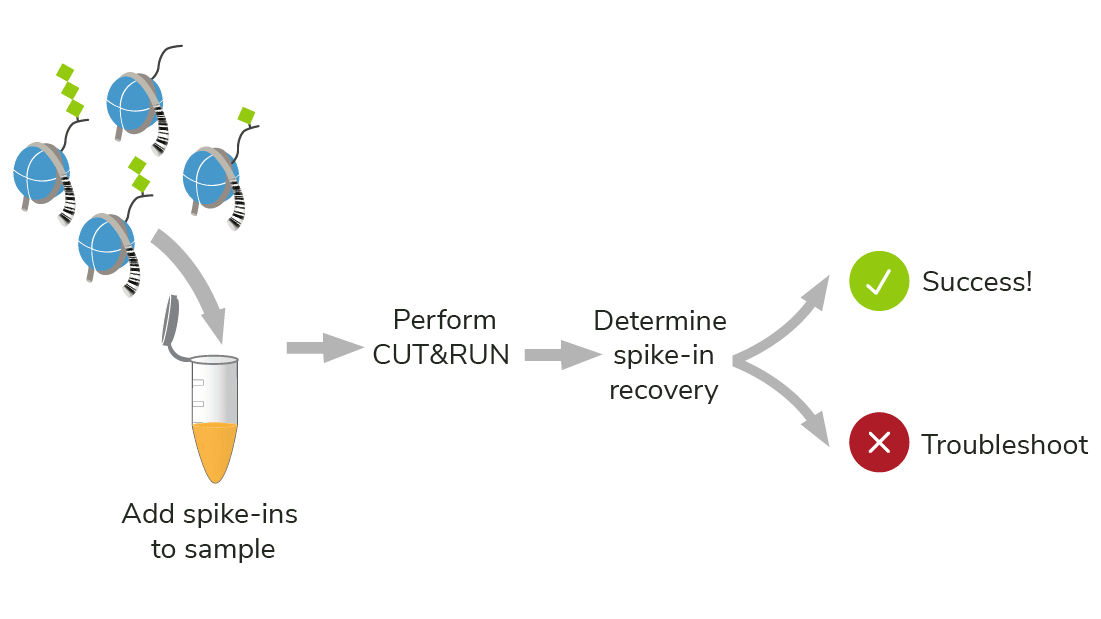
Exclusive spike-in control technology
Standard epigenomics controls can only validate part of your experiment. SNAP-CUTANA Spike-ins Controls use purified nucleosomes, which replicate the natural target of CUT&RUN, allowing you to:
- Directly monitor assay success
- Troubleshoot experiments
- Examine histone PTM antibody specificity
High-confidence epigenomic profiling
There are enough variables in experiments – let EpiCypher worry about the controls. Our CUTANA CUT&RUN kit and protocol include controls at every step, saving both time and resources.
- Quick methods to confirm quality sample prep
- Control reactions to monitor experimental success
- Clear metrics for confidence in proceeding to sequencing
Key Publications and Example Applications
Foundational CUT&RUN papers
Skene and Henikoff. An efficient targeted nuclease strategy for high-resolution mapping of
DNA binding
sites. eLife (2017). PMID 28079019.
This is the first paper describing CUT&RUN technology. Compared to ChIP-seq, the authors
show that CUT&RUN has enhanced sensitivity,
improved signal-to-noise, and reduced sequencing requirements.
Meers et al. Improved CUT&RUN chromatin profiling tools.
eLife (2019). PMID
31232687.
This is the original paper reporting the development of pAG-MNase, which forms the basis
of EpiCypher’s
CUTANA CUT&RUN technology.
CUTANA CUT&RUN in action
For histone PTMs
Chen et al. Distinct dynamics and functions of H2AK119ub1 and H3K27me3 in mouse preimplantation embryos. Nature Genetics (2021). PMID 33821005.
- CUT&RUN targets: H2AK119ub1, H3K27me3
- Sample types: Mouse germ cells and early-stage embryos
Yusufova et al. Histone H1 loss drives lymphoma by disrupting 3D chromatin architecture. Nature (2020). PMID 33299181.
- Targets: H3K9me2, H3K9me3
- Cell Types: Mouse germinal center B cells
Carpenter et al. Cell-type specific profiling of histone post-translational modifications in the adult mouse striatum. Nature Communications (2022). PMID 36513652.
- Targets: H3K4me3, H3K27me3
- Cell Types: Neuronal subtypes from mouse striatum
Wang et al. Systematic comparison of CRISPR-based transcriptional activators uncovers gene-regulatory features of enhancer–promoter interactions. Nucleic Acids Research (2022). PMID 35849129.
- Targets: H3K27ac
- Cell Types: Transfected HeLa and HEK293T cells
For chromatin-associated proteins
Zhu et al. Mitotic bookmarking by SWI/SNF subunits. Nature (2023). PMID 37225980.
- Targets: SOX2, ESRRB, SMARCE1, SMARCB1, SMARCA4, BRD9, ARID1A, EZH2
- Cell Types: Asynchronous and mitotic mouse embryonic stem cells
McDonald et al. Canonical BAF complex activity shapes the enhancer landscape that licenses CD8+ T cell effector and memory fates. Immunity (2023). PMID 37315534.
- Targets: ARID1A, H3K27ac, ETS1, BATF, T-bet
- Cell Types: Mouse CD8 T cells
Miao et al. Glucose dissociates DDX21 dimers to regulate mRNA splicing and tissue differentiation. Cell (2023). PMID 36608661.
- Targets: DDX21 and HA-tagged DDX21
- Cell Types: Primary human keratinocytes
Bomber et al. Human SMARCA5 is continuously required to maintain nucleosome spacing. Molecular Cell (2023). PMID 36630954.
- Targets: CTCF, Flag-tagged SMARCA5, H2A.Z, H3K27me3, RUNX1
- Cell Types: Human cell lines Kasumi-1, HEL, and OCI-LY1
For challenging sample types
Dai et al. Massively parallel knock-in engineering of human T cells. Nature Biotechnology (2023). PMID 36702900.
- Targets: PRDM1
- Cell Types: human CAR T cells
Sanchez-Priego et al. Mapping cis-regulatory elements in human neurons links psychiatric disease heritability and activity-regulated transcriptional programs. Cell Reports (2022). PMID 35649373.
- Targets: H3K27ac, FOS
- Cell Types: Human pluripotent stem cell-derived neurons
Larsen et al. Establishment, maintenance, and recall of inflammatory memory. Cell Stem Cell (2021). PMID 34320411.
- Targets: FOS, JUN, STAT3, ATF3, p63, Pol II, AP1
- Cell Types: FACS-purified mouse epidermal stem cells
Soto et al. Simplification of culture conditions and feeder-free expansion of bovine embryonic stem cells. Scientific Reports (2021). PMID 34040070.
- Targets: H3K4me3, H3K27me3
- Cell Types: Bovine embryonic stem cells
Michealraj et al. Metabolic regulation of the epigenome drives lethal infantile ependymoma. Cell (2020). PMID 32445698.
- Targets: H3K27me3
- Cell Types: Cultured human posterior fossa A (PFA) ependymomas
For a full list of publications citing EpiCypher’s products, click here.