CUTANA™ CUT&Tag Assays
Powerful assays for ultra-sensitive epigenomics
CUTANA™ CUT&Tag assays provide a streamlined, cost-effective workflow for chromatin mapping. These assays bypass the most challenging steps of traditional ChIP-seq profiling strategies, resulting in multiple advantages:
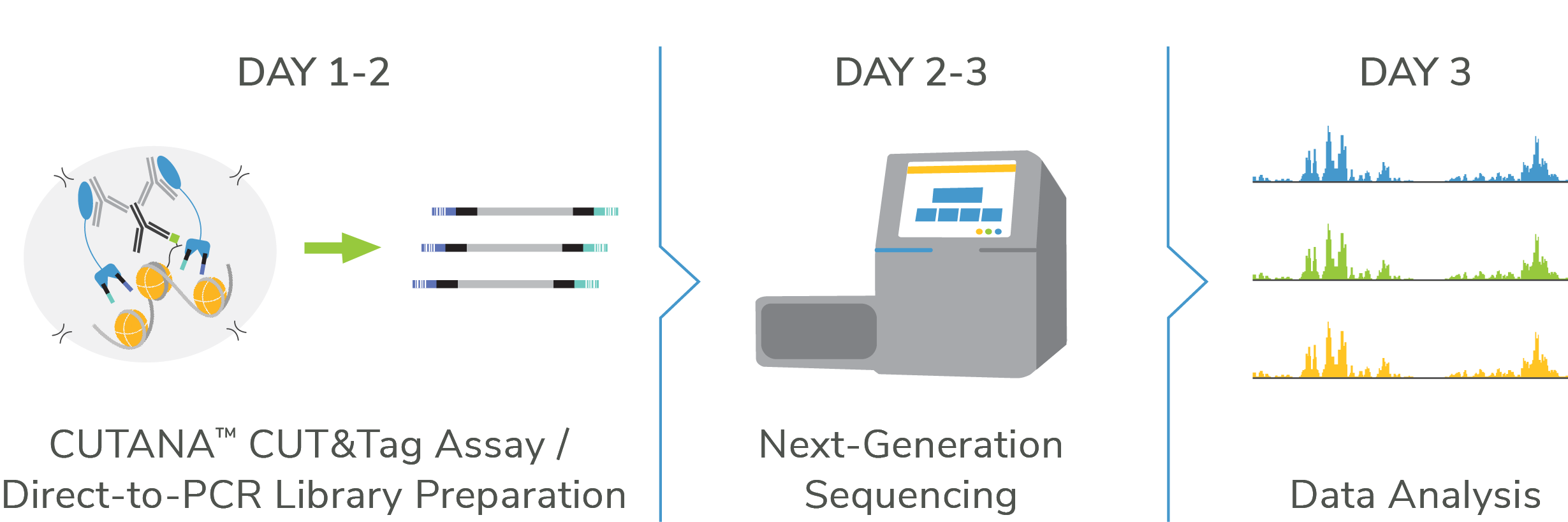
- Cells to libraries < 2 days using exclusive one-tube protocol
- High signal over background
- Reliable profiles for low cell numbers
- Cost-efficient sequencing with only 5-8 million reads per sample
- Robust assay kits, protocols, and controls available!
Have Questions?
We’re here to help. Click below and a member of our team will get back to you shortly!
CUTANA™ CUT&Tag provides impressive gains in histone PTM mapping compared to ChIP-seq, the traditional chromatin mapping assay.
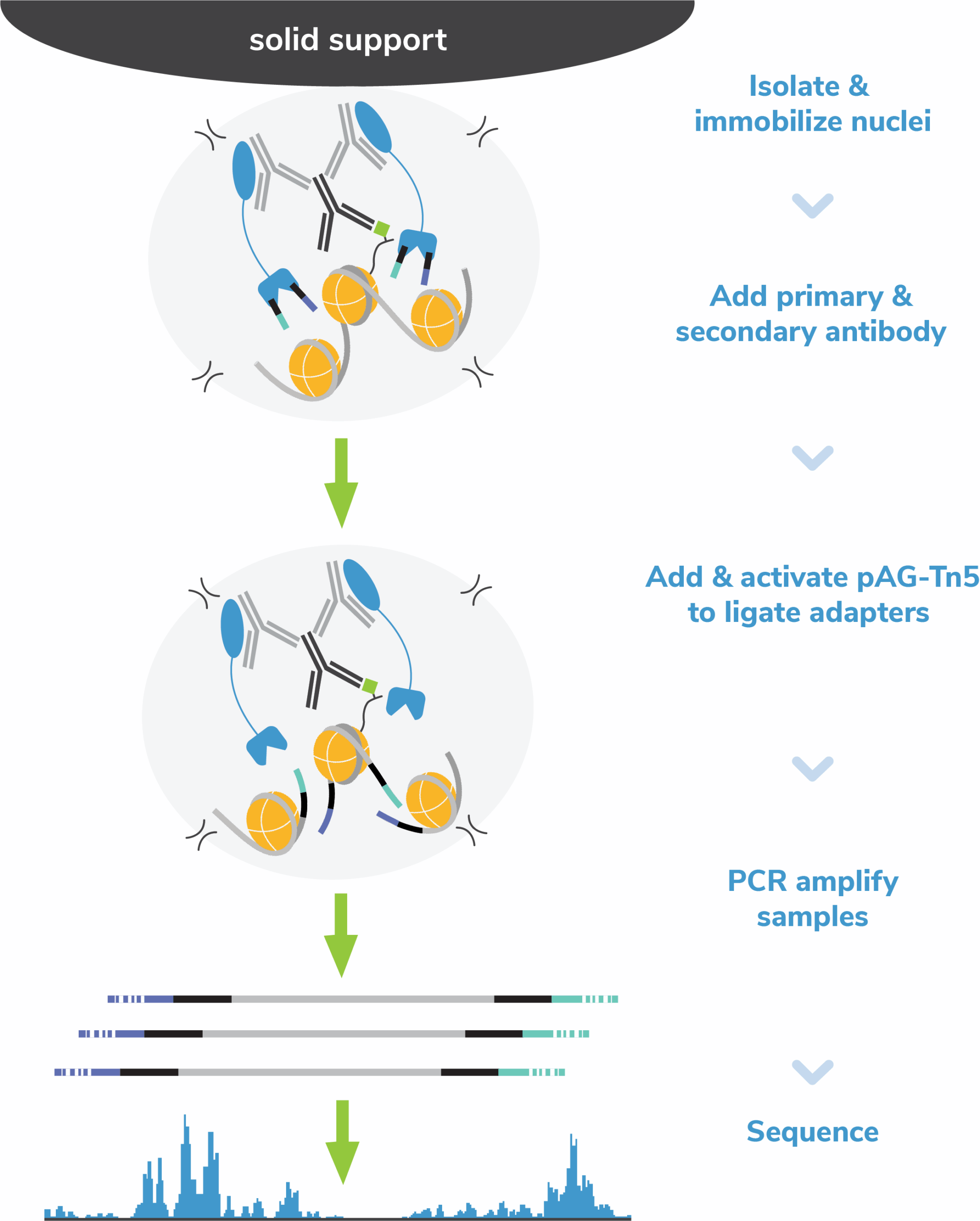
How does CUT&Tag work?
Cleavage Under Targets and Tagmentation (CUT&Tag) is a novel immunotethering-based chromatin mapping assay.
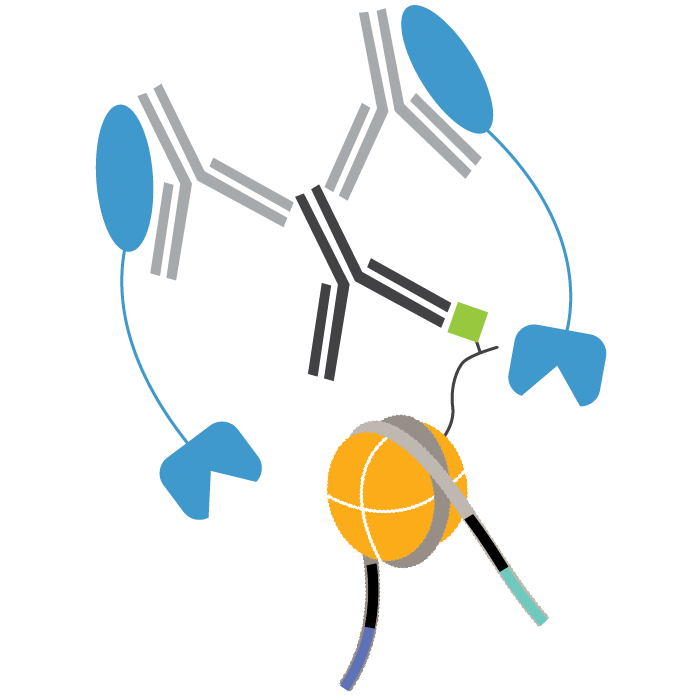
In CUT&Tag, a fusion of protein A, protein G, and Tn5 transposase (pAG-Tn5) is used to cleave and add sequencing adapters at antibody-bound chromatin. Tagmented DNA is selectively amplified by PCR and sequenced.
This strategy eliminates chromatin fragmentation, IP, and library prep, streamlining CUT&Tag workflows vs. ChIP-seq. Resulting profiles have improved signal over background using small numbers of cells and deliver major cost savings.
Is CUT&Tag right for your project? See this blog for help and try our CUT&Tag Kit to get started!
Get started with CUTANA™ CUT&Tag assays

CUTANA™ CUT&Tag Kit
Get the reagents you need for CUT&Tag in one convenient box. The kit also includes a comprehensive manual, with troubleshooting tips and FAQs.
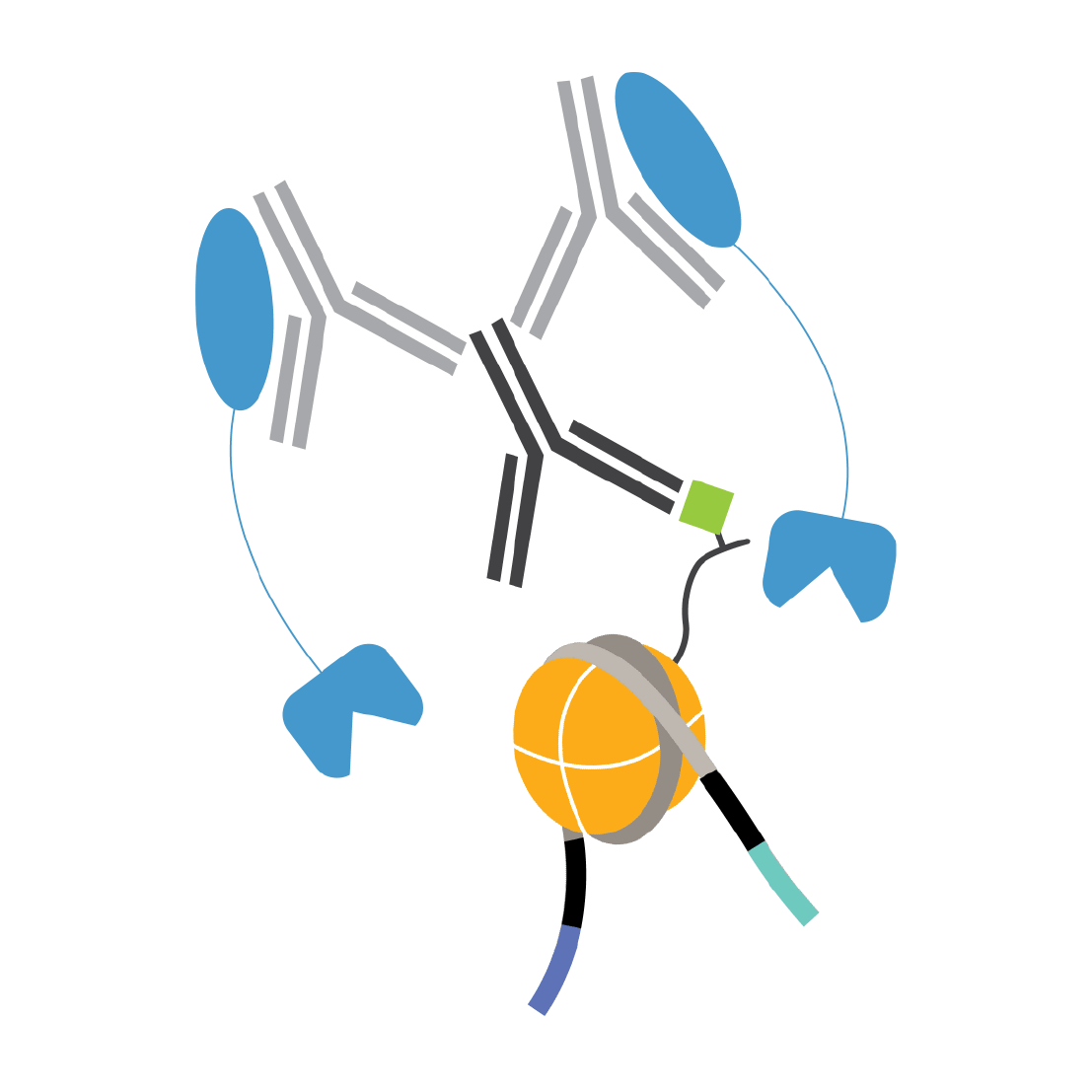
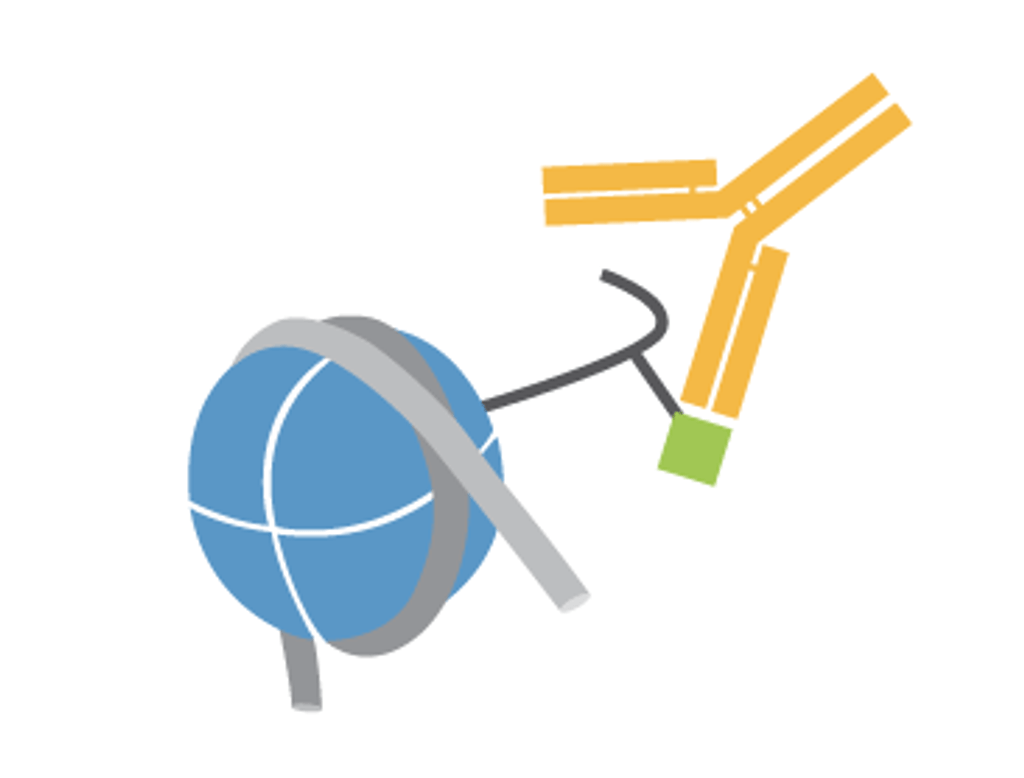
CUTANA™ pAG-Tn5 for CUT&Tag
Plan your next CUT&Tag experiment! pAG-Tn5 cleaves and appends sequencing adapters to antibody-bound chromatin.
CUTANA™ CUT&Tag Antibodies
Map with confidence. Our histone PTM antibodies are validated directly in CUT&Tag for high on-target specificity and efficiency.
The CUTANA™ CUT&Tag Protocol & Ideal Applications
EpiCypher makes it easy to customize your CUT&Tag experience by offering an all-inclusive kit, individual reagents & tools, and a user-friendly CUT&Tag protocol.
Both the CUT&Tag protocol and kit manual feature our rapid, one-tube strategy and extensive troubleshooting guidance. These resources are also routinely updated to reflect advances by EpiCypher scientists. The current protocol and kit are optimized for:
- Histone PTMs*
- Fresh, frozen, and cross-linked nuclei (preferred) or cells
- Low inputs (100,000 down to 10,000 starting cells)
- Reduced sequencing depths (5-8 million reads)
* We recommend CUTANA CUT&RUN assays to map chromatin-associated proteins. Trying to decide between CUT&RUN and CUT&Tag? Read our blog to learn more.
Explore Our Products
Featured Publications
- Kaya-Okur et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Comm. 10, 1930 (2019). (PMID: 31036827)
This paper introduces CUT&Tag methodology, as developed by the Henikoff laboratory at the Fred Hutchinson Cancer Research Center. It describes how the Tn5 fusion proteins works, demonstrates the application of CUT&Tag for histone PTM and chromatin factor profiling, and reveals major advantages of this new technology. Specifically, they use CUT&Tag to generate high-quality histone PTM profiles from single cells, a major advancement for the epigenetics field. - Kaya-Okur et al. Efficient low-cost chromatin profiling with CUT&Tag. Nat. Protoc. 15, 3264-3283 (2020). (PMID: 32913232)
Here, the inventors of CUT&Tag provide a highly detailed description of their optimized CUT&Tag protocol, citing EpiCypher’s pAG-Tn5 as the suggested commercial vendor of the enzyme. The protocol highlights critical steps, includes a troubleshooting FAQ table, and shows example results. - Henikoff et al. Efficient transcription-coupled chromatin accessibility mapping in situ. by nucleosome-tethered tagmentation. Elife 16;9:e63274.(2020). (PMID: 33191916)
The inventors of CUT&RUN / CUT&Tag technology leveraged EpiCypher’s pAG-Tn5 and CUT&RUN compatible histone PTM antibodies to develop a modified version of CUT&Tag for chromatin accessibility mapping. Importantly, this new method generates robust, high-quality profiles using only 3-5M reads/samples (vs. 20-30M reads for standard ATAC-seq). - Janssens et al. Automated CUT&Tag profiling of chromatin heterogeneity in mixed-lineage leukemia. Nat. Genet. 53, 1586-1596 (2021). (PMID: 34663924)
Assay automation is a major feature of CUT&Tag, as this will enable higher throughput for clinical research projects. In this study, Janssens et al. develop an automated CUT&Tag platform and use it to map oncofusion proteins, transcription factors, and histone PTMs in samples from leukemia patients. They also modified their approach for single cell analysis to assess inter- and intra-tumor heterogeneity. Janssens et al. CUT&Tag2for1: a modified method for simultaneous profiling of the accessible and silenced regulome in single cells Genome Biol. 23, 81.(2022). (PMID: 35300717)
CUT&Tag2For1 is an innovative application of CUT&Tag profiling technology that allows simultaneous, high-resolution mapping of transcription initiation (RNA Pol II phospho S5) and repressive domains (H3K27me3) in single cells or bulk populations. Mapping multiple chromatin features in a single reaction is important when cell numbers are limiting, including biopsy studies and clinical research.
To see a full list of publications, click here.
Related Blog Posts
- Aaron Alcala, PhD
Epigenetic mechanisms shape nearly every aspect of neurobiology—from guiding early neural development to regulating plasticity, cognition, and responses to injury...
- Aaron Alcala, PhD
Funding challenges are a reality for many researchers, and we want to make sure innovation doesn’t slow down because of...
- Hannah Devens, PHD
CUT&RUN and CUT&Tag are powerful chromatin mapping tools driving innovative research. Over the years, we have written comprehensive blog posts...
- Hannah Devens, PHD
CUT&RUN and CUT&Tag protocols have transformed the epigenetics field by offering faster, more accessible alternatives to traditional ChIP-seq chromatin mapping...

- Ellen Weinzapfel
Multiomic studies, which involve the integration of DNA, RNA, and protein “omics” datasets from a single cell or sample type,...

- Ellen Weinzapfel
Purification of target-enriched DNA is central to CUT&RUN (and CUT&Tag) chromatin mapping workflows. When considering a DNA cleanup strategy, multiple...