CUTANA™ ChIC/CUT&RUN Kit
Save 10% when you buy with our Library Prep Kit for a streamlined CUT&RUN workflow!
The CUTANA™ ChIC/CUT&RUN Kit enables streamlined chromatin profiling of histone post-translational modifications (PTMs) and chromatin associated proteins.
Key Advantages- User-friendly – The kit includes all reagents needed to go from starting cells to isolated DNA in a robust and scalable multi-channel workflow.
- Integrated workflow solution – Part of our comprehensive CUTANA™ ecosystem, this kit pairs seamlessly with the CUTANA™ CUT&RUN Library Prep Kit (14-1001) and CUTANA™ Cloud to accelerate your path from cells to insights.
- Ideal for diverse applications – Kit protocol is optimized for use with a variety of chromatin targets, starting cell numbers, and sample preparation methods.
- Controls drive reliability – Included control antibodies and defined spike-ins ensure results you can trust.
- Ready to run, fully supported – Every kit comes with a manual packed with detailed protocols, quality checks, FAQs, and expert tips, plus a bench-proof Quick-Start Card and access to the Tech Support Center for additional guidance.
- Stringent quality control – Each new lot undergoes rigorous quality control checks to ensure consistent results in your research.
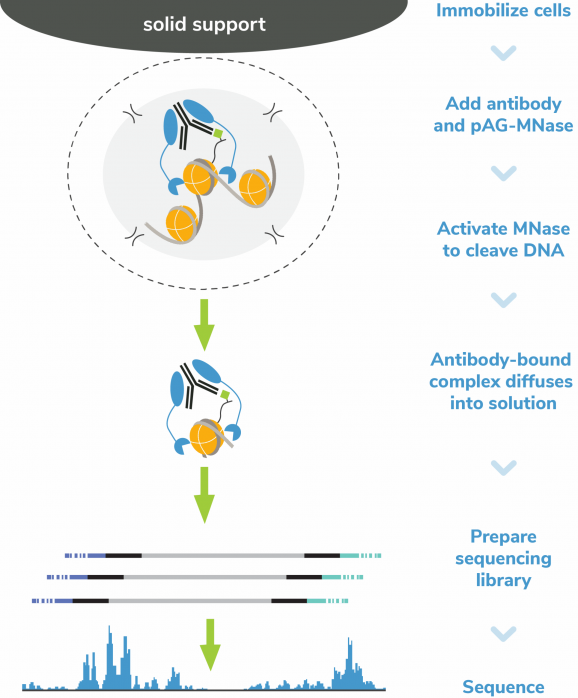
In CUT&RUN, pAG-MNase selectively releases antibody-bound chromatin into solution, where it is easily separated from bead-bound cells and purified to create high quality libraries for next-generation sequencing. This results in high sensitivity chromatin mapping using small numbers of starting cells and low sequencing depth (5-10 million total reads). The CUTANA™ ChIC/CUT&RUN Kit is optimized for use with a variety of targets including histone PTMs, transcription factors, chromatin remodelers, epigenetic enzymes, and reader proteins. The kit is compatible with diverse samples including cells or nuclei derived from native, cryopreserved, or cross-linked samples. While it is recommended to start with 500,000 cells, comparable data can be generated using as few as 5,000 cells.
The CUT&RUN Kit Version 6 (v6) now includes two new Wash Buffer Enhancers to reduce clumping and improve bead handling and CUTANA™ DNA Purification Beads with an updated bead-to-sample ratio to preserve precise size selection and improve capture of smaller transcription factor-associated fragments. Each kit is equipped with a comprehensive selection of controls to ensure reliable results: positive (H3K4me3 and H3K27me3) and negative (IgG) control antibodies paired with SNAP-CUTANA™ spike-ins (Figure 2) support assay optimization and continuous monitoring, while E. coli Spike-in DNA enables data normalization. The kit utilizes multi-channel pipetting with provided 8-strip tubes to ensure seamless handling while maximizing throughput and reproducibility. CUTANA™ ChIC/CUT&RUN Kits are bench-tested, scientist-approved, providing users with quality reagents for precision mapping.
To place bulk orders, contact us. Keep learning by checking out our Tech Support Center.
Intellectual Property
US Pat. No. 7790379, 11885814 and related patents and pending applications.
Beckman Coulter, the stylized logo, and SPRIselect are trademarks or registered trademarks of Beckman Coulter, Inc. in the United States and other countries.
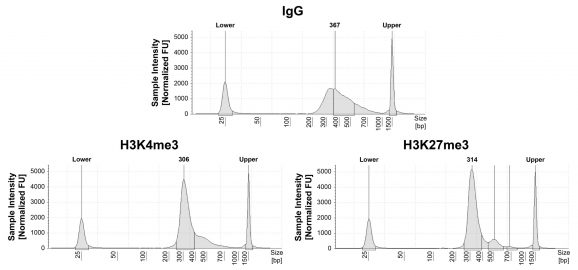
Figure 1: CUT&RUN DNA fragment size distribution analysis
CUT&RUN was performed as described in Figure 5. Library DNA was analyzed by Agilent Tapestation®. This analysis confirmed that mononucleosomes were predominantly enriched in CUT&RUN (~300 bp peaks represent 150 bp nucleosomes + sequencing adapters).
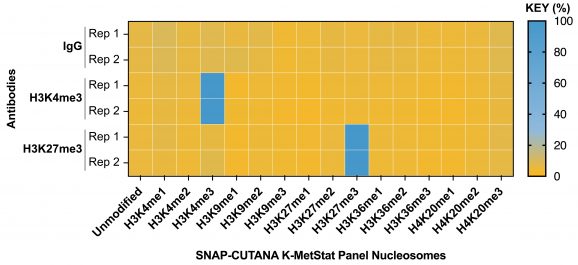
Figure 2: SNAP-CUTANA™ Spike-in controls
DNA-barcoded designer nucleosomes (dNucs) harboring distinct K-methyl PTMs were spiked into CUT&RUN reactions prior to antibody addition. Spike-in barcodes were analyzed using the shell script available on the spike-in product page (EpiCypher 19-1002). Barcodes for IgG (normalized to total reads), H3K4me3 and H3K27me3 (normalized to on-target) antibodies are shown. The spike-ins confirmed H3K4me3 and H3K27me3 antibodies specifically recovered the target dNucs, while IgG showed no preferential enrichment.
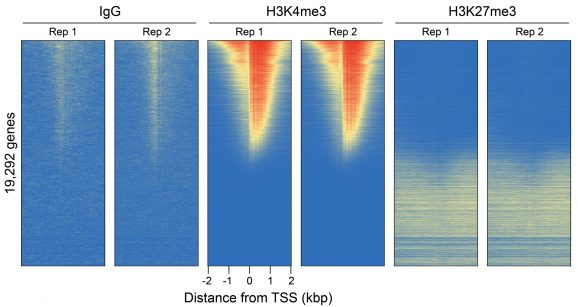
Figure 3: CUT&RUN genome-wide heatmaps
CUT&RUN was performed as described in Figure 5. Heatmaps show two replicates (“Rep”) of IgG, H3K4me3, and H3K27me3 kit control antibodies in aligned rows ranked by intensity (top to bottom) relative to the H3K4me3 Rep 1 reaction and colored such that red indicates high localized enrichment and blue denotes background signal.
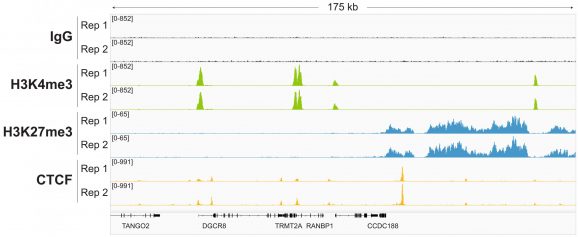
Figure 4: Representative gene browser tracks
CUT&RUN was performed as described in Figure 5. A representative 175 kb window at the TRMT2A gene is shown for two replicates (“Rep”) of IgG, H3K4me3, and H3K27me3 kit control antibodies. Representative tracks are also shown for the transcription factor CTCF. The CUT&RUN kit produced the expected genomic distribution for each target. Images were generated using the Integrative Genomics Viewer (IGV, Broad Institute).
Figure 5: CUT&RUN methods
CUT&RUN was performed using the CUTANA™ ChIC/CUT&RUN Kit starting with 500k K562 cells with 0.5 µg of IgG (EpiCypher 13-0042), H3K4me3 (EpiCypher 13-0060), H3K27me3 (EpiCypher 13-0055), or 0.125 µg of CTCF (EpiCypher 13-2014) antibodies in duplicate. Library preparation was performed with 5 ng of DNA (or the total amount recovered if less than 5 ng) using the CUTANA™ CUT&RUN Library Prep Kit (EpiCypher 14-1001/14-1002). Libraries were run on an Illumina NextSeq2000 with paired-end sequencing (2×50 bp). Sample sequencing depth was 5.5/18.8 million reads (IgG Rep 1/Rep 2), 14.2/17.0 million reads (H3K4me3 Rep 1/Rep 2), 24.7/18.1 million reads (H3K27me3 Rep 1/Rep 2), and 8.6/5.5 million reads (CTCF Rep 1/Rep 2). Data were aligned to the T2T-CHM13v2.0 genome using Bowtie2. Data were filtered to remove duplicates, multi-aligned reads, and ENCODE DAC Exclusion List regions.
Storage
OPEN KIT IMMEDIATELY and store components at room temperature, 4°C, and -20°C as indicated (see User Manual corresponding to Kit Version 6). Stable for 12 months upon date of receipt.
Instructions for Use
See User Manual corresponding to Kit Version 6. This kit is not compatible with previous user manuals.
| Item | Cat. No. |
|---|---|
| CUTANA™ pAG-MNase for ChIC/CUT&RUN Workflows | 15-1016k |
| H3K4me3 Antibody, SNAP-Certified™ for CUT&RUN | 13-0060k |
| H3K27me3 Antibody, SNAP-Certified™ for CUT&RUN | 13-0055k |
| CUTANA™ Rabbit IgG CUT&RUN Negative Control Antibody | 13-0042k |
| SNAP-CUTANA™ K-MetStat Panel | 19-1002k |
| CUTANA™ Concanavalin A Conjugated Paramagnetic Beads | 21-1401k |
| CUTANA™ DNA Purification Beads | 21-1407k |
| CUTANA™ Wash Buffer Enhancer 1 | 21-1028k |
| CUTANA™ Wash Buffer Enhancer 2 | 15-1030k |
| CUTANA™ E. coli Spike-in DNA | 18-1401k |
| CUTANA™ CUT&RUN 8-Strip 0.2 mL Tubes | 10-0009k |
| Bead Activation Buffer | 21-1001k |
| Pre-Wash Buffer | 21-1002k |
| Stop Buffer | 21-1003k |
| 5% Digitonin | 21-1004k |
| 1 M Spermidine | 21-1005k |
| 0.5 M EDTA | 21-1006k |
| 100 mM Calcium Chloride | 21-1007k |
| 0.1X TE Buffer | 21-1025k |
| Item | Cat. No. |
|---|---|
| CUTANA™ CUT&RUN Library Prep Kit | 14-1001/14-1002 |
| SNAP-CUTANA™ K-MetStat Panel | 19-1002 |
| CUT&RUN Antibodies | See the list |
| Magnetic Separation Rack, 0.2 mL Tubes | 10-0008 |
| Magnetic Separation Rack, 1.5 mL Tubes | 10-0012 |
| CUTANA™ Nuclei Extraction Buffer | 21-1026 |
| CUTANA™ Protease Inhibitor Tablets | 21-1027 |
| CUTANA™ DNA Purification Beads | 21-1407 |