High Sensitivity. Rapid Results.
Superior Chromatin Mapping.
CUTANA™ Assays for CUT&RUN and CUT&Tag are transforming epigenomics. These highly sensitive workflows enable the study of low cell numbers and challenging targets, expanding chromatin mapping capabilities for clinical research and precious samples. Compared to ChIP-seq, the traditional profiling assay, CUTANA assays offer multiple advantages:
- Fewer cells required
- Profile a wider range of targets
- Simplified workflow with no fragmentation or IP
- Dramatically reduced costs
- Robust workflows optimized by EpiCypher scientists
Have Questions?
We’re here to help. Click below and a member of our team will get back to you shortly!
Request More Info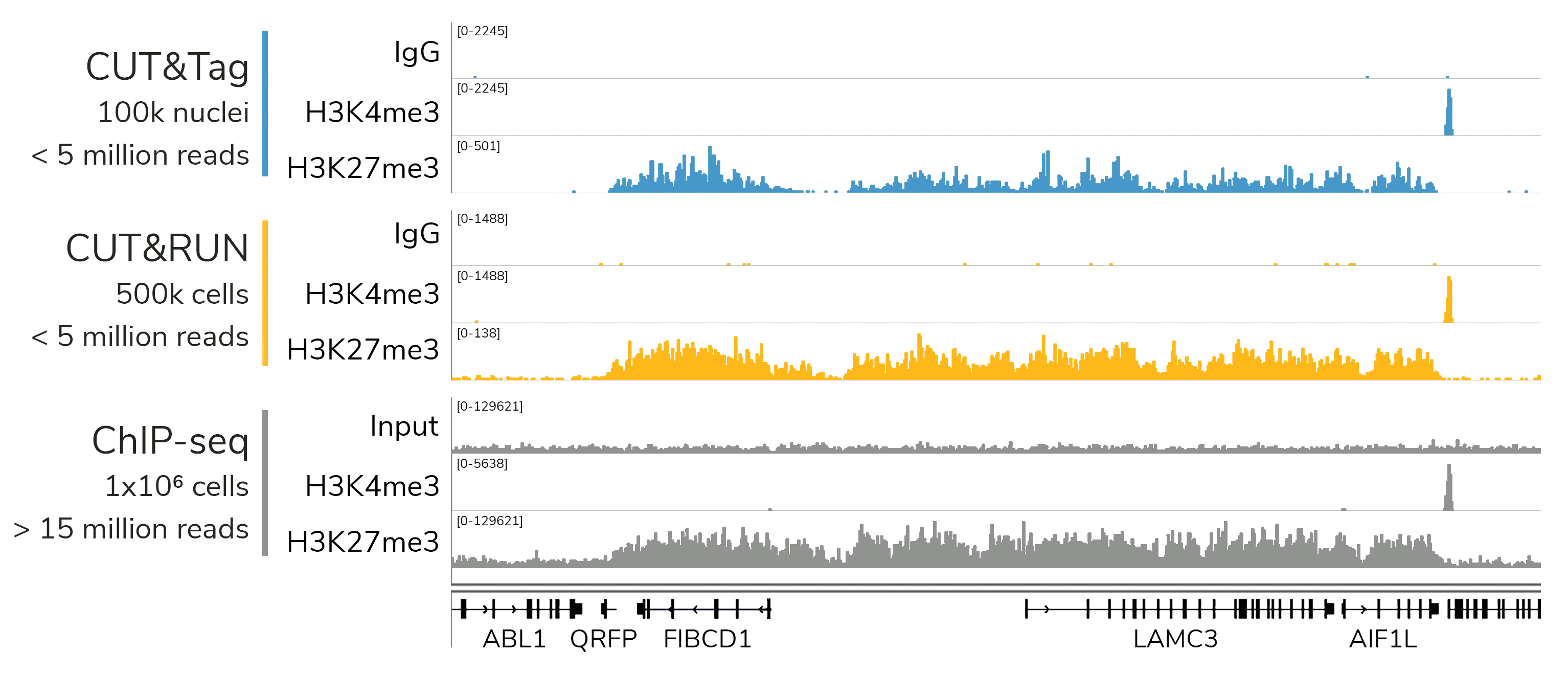
CUTANA™ assays generate high-quality epigenomic profiles with a fraction of the cells and sequencing depth needed for ChIP-seq. A representative 300 kb region is shown for CUT&Tag (blue), CUT&RUN (orange), and ChIP-seq (gray).
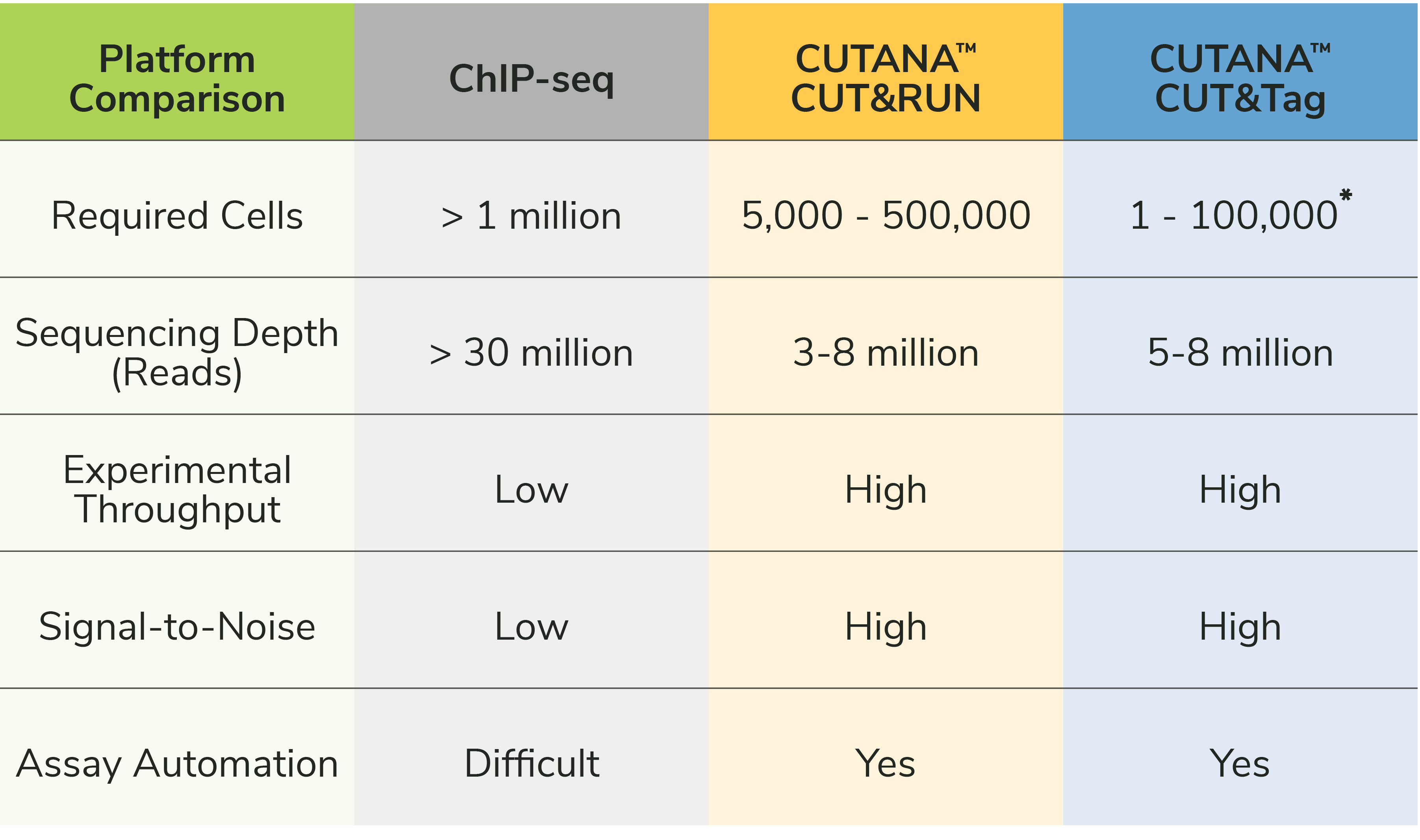
CUTANA™ Assays vs. ChIP-seq:
Streamlined workflows, faster results, higher quality data

Get the details: A platform-by-platform comparison
CUT&Tag has been applied for single-cell chromatin analysis in the literature (see papers below). However, the recommended input for CUTANA™ CUT&Tag assays is 10,000 to 100,000 cells.
CUT&RUN vs CUT&Tag: Which is right for you?
CUT&RUN: EpiCypher's go-to epigenomics assay
- A flexible assay for broad applications
- Diverse targets and cell types
- User-friendly workflow
- Robust profiles with high signal-to-noise
CUTANA™ assays are driving epigenetics forward
Researchers around the globe are leveraging CUTANA CUT&RUN and CUT&Tag assays to make exciting discoveries in the epigenetics field. See what some of our customers have been up to!
CUTANA™ CUT&RUN
- Yusufova et al. Histone H1 loss drives lymphoma by disrupting 3D chromatin architecture. Nature 589, 299–305 (2021). (PMID: 33299181)
- Yuan et al. Elevated NSD3 histone methylation activity drives squamous cell lung cancer. Nature 590, 504–508 (2021). (PMID: 33536620)
- Patty & Hainer. Transcription factor chromatin profiling genome-wide using uliCUT&RUN in single cells and individual blastocysts. Nat. Protoc 16, 2633-2666 (2021). (PMID: 33911257)
- Larson et al. Establishment, maintenance, and recall of inflammatory memory. Cell Stem Cell. 28, 1758-1774 (2021). (PMID: 34320411)
CUTANA™ CUT&Tag
- Henikoff et al. Efficient chromatin accessibility mapping in situ by nucleosome-tethered tagmentation. eLife 16;9:e63274 (2020). (PMID: 33191916)
- Kaya-Okur et al. Efficient low-cost chromatin profiling with CUT&Tag. Nat. Protoc. 15, 3264-3283 (2020). (PMID: 32913232)
- Kaya-Okur et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Comm. 10, 1930 (2019). (PMID: 31036827)
- Janssens et al. CUT&Tag2for1: a modified method for simultaneous profiling of the accessible and silenced regulome in single cells. Genome Biol. 23, 81 (2022). (PMID: 31036827)
- Janssens et al. Automated CUT&Tag profiling of chromatin heterogeneity in mixed-lineage leukemia. Nat. Genet. 53, 1586-1596 (2021). (PMID: 34663924)

