CUT&RUN vs. CUT&Tag : Which One is Right for You?

In recent years, the fields of genomics and epigenetics have been introduced to two exciting and novel immunotethering approaches for chromatin profiling: CUT&RUN (Cleavage Under Targets and Release Using Nuclease)1-4 and CUT&Tag (Cleavage Under Targets and Tagmentation)5.
The hype surrounding these technologies is driven by their ability to overcome many of the limitations and drawbacks associated with classical chromatin mapping approaches, like ChIP-seq (Chromatin ImmunoPrecipitation sequencing). Indeed, CUT&RUN and CUT&Tag provide streamlined chromatin mapping using fewer cells and lower sequencing depths, thus generating higher quality data at a reduced price point.
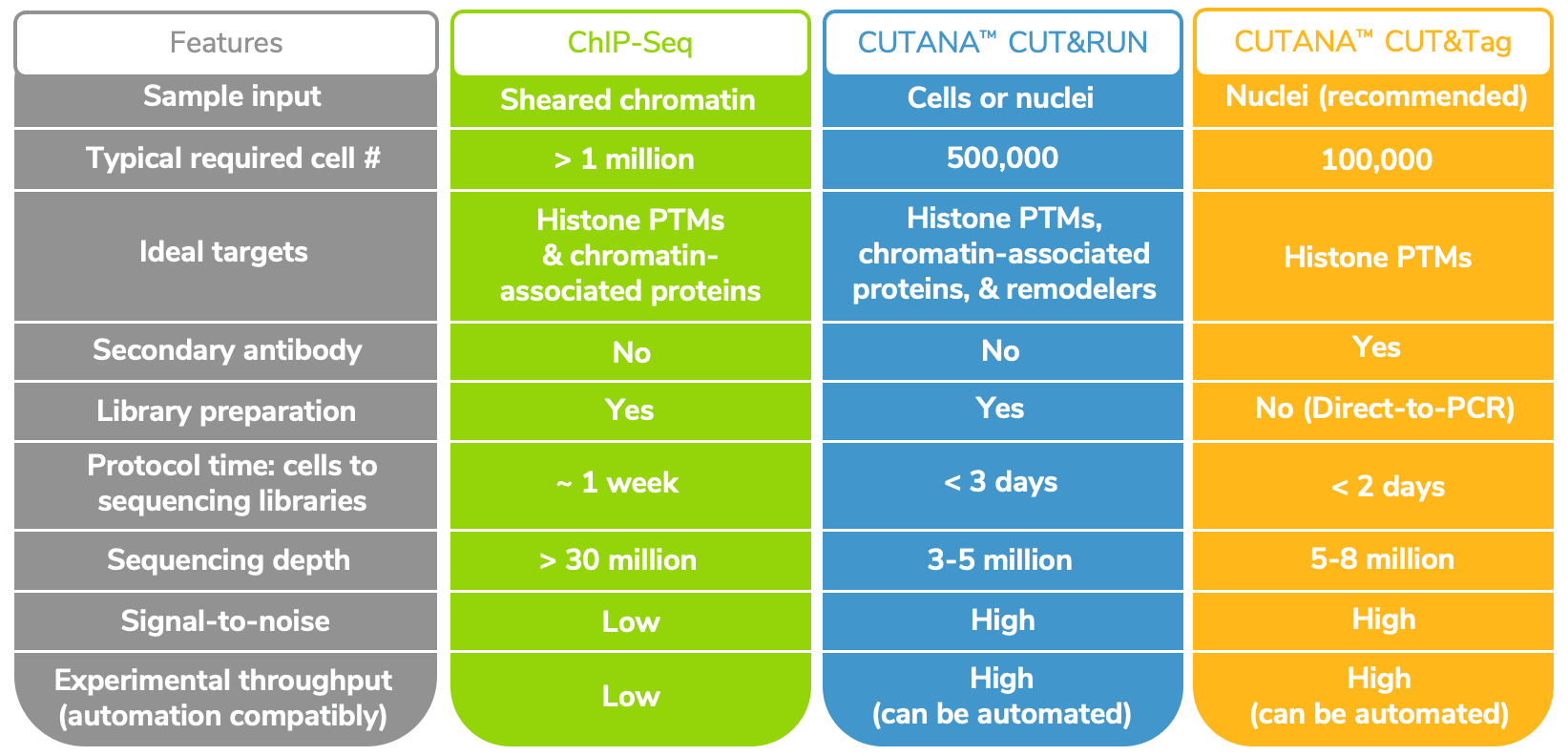
Table 1 below summarizes the major distinctions across both platforms in comparison to ChIP-seq using EpiCypher CUTANATM CUT&RUN and CUT&Tag assays.
Questions on approach
Many first-time users are confused as to which assay will best fit their needs. Some of the most common questions include:
- Since CUT&Tag bypasses library preparation, I should always use CUT&Tag, right?
- Are there situations in which CUT&RUN is actually the preferred method? Why?
In this post we will be diving into the nuances of each method to help researchers determine the best technical approach for their biological question of interest.
Immunotethering basics
We’ve covered each assay in depth on our blog (click here for CUT&RUN and here for CUT&Tag). Briefly, both CUT&RUN and CUT&Tag leverage a protein A/G (pAG) fusion tag to “tether” an enzyme to antibody-labelled chromatin loci for directed cleavage.
In the case of CUT&RUN, pAG is fused to micrococcal nuclease (pAG-MNase), and chromatin fragmentation at antibody-bound loci is activated by addition of calcium2-4. In CUT&Tag, pAG is fused to a hyperactive Tn5 transposase (pAG-Tn5) pre-loaded with sequencing adapters, and is activated by magnesium to simultaneously fragment and “tag” antibody-labelled chromatin with adapters5.
EpiCypher’s CUTANA CUT&RUN and CUT&Tag protocols are optimized for 500,000 and 100,000 cells, respectively, although both immunotethering assays have been adapted for single cell inputs by other labs5, 6. Our CUT&RUN protocol produces robust mapping data for histone post-translational modifications (PTMs) using as little as 5,000 cells without any change to the protocol.
The straightforward workflows for CUT&RUN and CUT&Tag allow users to go from cells to sequencing libraries in approximately 2 days (see Table 1). Furthermore, CUT&Tag and CUT&RUN only require 3-8 million reads per sample, compared to the 30 million (or more) reads needed for equivalent coverage by ChIP-seq. Despite the reduced sequencing depths, these new approaches have improved signal-to-noise and substantially reduced backgrounds vs. ChIP-seq.
These remarkable gains in sensitivity mean that more experiments can be performed at a lower cost. It also broadens the application of benchtop sequencers (e.g. Illumina Miniseq and MiSeq), lower-cost alternatives to the high-throughput machines required for ChIP-seq projects (e.g. NextSeq). Benchtop sequencers generate 30-40 million reads per run, and thus cannot be used for ChIP-seq projects, but are perfect for rapid, multiplexed sequencing of up to 10 CUT&RUN or CUT&Tag samples.
Advantages of CUT&Tag : Streamlined sample processing
Taken at face value, both assays are fantastic! But there are several features of CUT&Tag that make it an attractive method to chromatin scientists.
- Access to low and single-cell inputs. Single cell profiling is a rapidly growing area of biomedical research7, but it has been challenging to profile chromatin from these low inputs. EpiCypher's CUT&Tag protocol and CUT&Tag Kitgenerate quality data down to 10,000 cells without adjusting the workflow. With unique adaptations, CUT&Tag has been used to map histone PTMs from ultra-low nuclear inputs (down to 60 cells) and single-cells1, 5 which would increase access to heterogeneous cell populations in tumors/tissues and precious clinical samples.
- No library prep → save money. Sequencing library preparation is time consuming and expensive. The application of Tn5 for directed ligation of sequencing adapters in CUT&Tag bypasses traditional library prep steps (e.g. end polishing) for accelerated sample processing and significantly reduced assay costs5.
- Simplified workflow. As mentioned above, CUT&Tag enables rapid chromatin profiling, going from cells to next-generation sequencing libraries in only two days (five hours of hands-on work with overnight incubation step). Our exclusive Direct-to-PCR CUT&Tag method allows selective amplification of tagmented DNA from bulk nuclei (as opposed to purified DNA), enabling the entire workflow to be performed in a single tube, which further streamlines the assay1.
The efficient workflow, combined with the reduced number of steps, supports the use of CUT&Tag to profile single cells. Although CUT&RUN has been optimized for single cell analysis6, it requires significant modifications to the original protocol. CUT&Tag, however, was applied to single cells in its first publication5, highlighting the potential of this technology in single cell sequencing.
Click Here for Technical Support on CUT&RUN and CUT&Tag Assays!
Weaknesses of CUT&Tag: Target compatibility
With all of these benefits, it seems like CUT&Tag would be the preferred method, right?
But, despite the aforementioned advancements, CUT&Tag has some significant limitations.
- Off-target Tn5 activity and sample processing. CUT&Tag data may exhibit high backgrounds, reflecting Tn5 binding open chromatin and/or mitochondrial DNA (as in ATAC-seq). To avoid nonspecific signal from mitochondrial DNA, it is strongly recommended to use nuclei as opposed to whole cells. Thus, CUT&Tag may require optimization to successfully harvest intact nuclei from the desired cell population. Notably, CUT&RUN can be used to profile both cells and nuclei. So, if the type of cell or tissue you are working with is particularly challenging to dissociate, or if the nuclei are fragile, it may be best to try CUT&RUN.
-
Current protocols do not work for all target classes.
CUT&Tag generates robust data for histone PTMs, but is unreliable for
mapping chromatin associated proteins. Similarly, EpiCypher’s CUTANA CUT&Tag
protocol and CUT&Tag Kit are rigorously optimized for histone PTMs, but is
not recommended for chromatin-associated proteins.
In contrast, EpiCypher’s CUT&RUN protocol and CUT&RUN Kit are validated for histone PTMs, epigenetic reader proteins and enzymes, transcription factors, and even chromatin remodeling enzymes (notoriously challenging to profile by ChIP-seq). The reasons for this disparity between CUT&RUN and CUT&Tag methods are not fully understood and are a current topic of investigation at EpiCypher.
One potential factor is the difference in salt concentration between the two assays. CUT&Tag requires high-salt washes to remove non-specific/unbound pAG-Tn5 prior to tagmentation. This limited flexibility with salt concentrations may impact CUT&Tag data quality for weaker chromatin interactions. Indeed, updated CUT&RUN protocols have switched to a low-salt buffer, which improves sample recovery and sequencing resolution for many targets.
EpiCypher is continuing to optimize CUT&Tag fixation conditions, to determine if certain classes of proteins benefit from alternate cross-linking methods. Be sure to follow us on Twitter and sign up for emails to learn about new protocols!
CUTANATM CUT&Tag Kits Now Available!
Which method should I use for my target?
Regardless of target, we recommend trying CUT&RUN first, since this is a more robust assay (see Figure 1) and will give you reliable results for comparison to CUT&Tag. In addition, CUT&RUN is compatible with diverse cell types, fixation conditions, and targets, and can resolve DNA binding motifs with base pair resolution.

We recommend CUT&Tag for select applications, including histone PTM profiling from native (unfixed) cell populations and studies in which many samples need to be processed in a time- and cost-effective manner. Single cell assay development is also a potential application for CUT&Tag, but has yet to be developed and rigorously tested at EpiCypher. CUT&Tag is definitely for the more seasoned researcher, who is comfortable with epigenomic mapping approaches.
For both assays, we suggest performing your first experiment with the optimal input (see Table 1), and to always include positive (i.e. H3K4me3 or H3K27me3) and negative (i.e. anti-Rabbit IgG) controls. EpiCypher offers all the reagents you need to get started, including enzymes and reliable antibodies for assay controls.
You can always reach out to EpiCypher for help with setting up your CUT&RUN and/or CUT&Tag experiments, and for advice on which assay to use!
References
- Henikoff S et al. Profiling the epigenome at home. bioRxiv, (2020)
- Meers MP et al. Improved CUT&RUN chromatin profiling tools. Elife 8, (2019). PMID:31232687
- Skene PJ et al. Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat Protoc 13, 1006-19 (2018). PMID:29651053
- Skene PJ et al. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. Elife 6, (2017). PMID:28079019
- Kaya-Okur HS et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat Commun 10, 1930 (2019). PMID:31036827
- Hainer SJ et al. Profiling of Pluripotency Factors in Single Cells and Early Embryos. Cell 177, 1319-29 e11 (2019). PMID:30955888
- Method of the Year 2019: Single-cell multimodal omics. Nat Methods 17, 1 (2020). PMID:31907477
