CUTANA™ Fluorescent pAG-Tn5 for CUT&Tag
{"url":"https://www.epicypher.com/products/epigenetics-kits-and-reagents/cutana-cut-tag-assays/cutana-fluorescent-pag-tn5-for-cut-tag","add_this":[{"service":"facebook","annotation":""},{"service":"email","annotation":""},{"service":"print","annotation":""},{"service":"twitter","annotation":""},{"service":"linkedin","annotation":""}],"gtin":null,"id":934,"bulk_discount_rates":[],"can_purchase":true,"meta_description":"Fluorescently labeled pAG-Tn5 enables multiomic visualization and mapping of chromatin features in CUT&Tag.","category":["Epigenetics Kits and Reagents","Epigenetics Kits and Reagents/CUTANA™ CUT&Tag Assays","Other/IDEA Toolbox"],"AddThisServiceButtonMeta":"","main_image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/934/1048/toolbox-icon__95574.1661116173.png?c=2","alt":"CUTANA™ Fluorescent pAG-Tn5 for CUT&Tag"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=934","shipping":{"calculated":true},"num_reviews":0,"weight":"0.01 LBS","custom_fields":[{"id":"1152","name":"Pack Size","value":"125 µL"},{"id":"1153","name":"Internal Comment","value":"extra boxes in bottom shelf of Venom"}],"sku":"15-1026","description":"<div class=\"product-general-info\">\n <ul class=\"product-general-info__list-left\">\n <li class=\"product-general-info__list-item\">\n <strong>Type: </strong>Transposase\n </li>\n <li class=\"product-general-info__list-item\">\n <strong>Mol Wgt: </strong>192 kDa\n </li>\n </ul>\n <ul class=\"product-general-info__list-right\">\n <li class=\"product-general-info__list-item\">\n <strong>Host: </strong><em>E. coli</em>\n </li>\n <li class=\"product-general-info__list-item\">\n <strong>Epitope Tag: </strong>None\n </li>\n </ul>\n</div>\n<div class=\"service_accordion product-droppdown\">\n <div class=\"container\">\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel current\">\n <h3 class=\"sub-title1\">Description</h3>\n <div\n class=\"ProductDescriptionContainer product-droppdown__section-description-specific\"\n style=\"display: block\">\n <p>\n Products in EpiCypher’s IDEA Toolbox (Innovation and Discovery of\n Epigenetic Applications) offer access to reagents without known or\n fully defined uses, enabling researchers to explore cutting-edge\n applications. Due to their novelty and unexplored potential,\n EpiCypher will engage in limited technical support.\n </p>\n <p>\n CUTANA Fluorescent pAG-Tn5 is a fusion of Proteins A and G to\n <em>E. coli</em> transposase (Tn5), the key enzyme for CUT&Tag [1].\n The Mosaic A and Mosaic B adapters loaded in this enzyme bear a 5’\n Cy5 fluorescent dye to enable multiomic visualization and mapping of\n chromatin features. This product is highly purified to remove\n contaminating <em>E. coli</em> DNA. For an enzyme charged with\n non-fluorescent adapters that can be used in standard CUT&Tag, see\n EpiCypher\n <a\n href=\"https://www.epicypher.com/products/epigenetics-kits-and-reagents/cutana-pag-tn5-for-cut-and-tag\"\n >15-1017</a\n >.\n </p>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel current\">\n <h3 class=\"sub-title1\">Validation Data</h3>\n <div\n class=\"ProductDescriptionContainer product-droppdown__section-description-specific\"\n style=\"display: block\">\n <section class=\"image-picker\">\n <div class=\"image-picker__left\">\n <div\n class=\"image-picker__main-content_active image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg\n class=\"image-picker__svg-left\"\n width=\"24\"\n height=\"24\"\n viewBox=\"0 0 24 24\">\n <path\n d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a\n href=\"/content/images/products/proteins/15-1026-charging-data.jpeg\"\n target=\"blank\"\n class=\"image-picker__main-image-link\"\n ><img\n alt=\"15-1026-charging-data\"\n src=\"/content/images/products/proteins/15-1026-charging-data.jpeg\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\"\n >(Click to enlarge)</span\n ></a\n >\n <button class=\"image-picker__right-arrow\">\n <svg\n class=\"image-picker__svg-right\"\n width=\"24\"\n height=\"24\"\n viewBox=\"0 0 24 24\">\n <path\n d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\"\n ><strong>Figure 1: DNA charging data </strong><br />\n CUTANA Fluorescent pAG-Tn5 for CUT&Tag (0.5 µg) was resolved\n via Native PAGE alongside uncharged pAG-Tn5 (EpiCypher\n <a\n href=\"/products/epigenetics-kits-and-reagents/cutana-cut-tag-assays/cutana-uncharged-pag-tn5-for-cut-tag\"\n >15-1025</a\n >) and charged pAG-Tn5 (EpiCypher\n <a\n href=\"https://www.epicypher.com/products/epigenetics-kits-and-reagents/cutana-pag-tn5-for-cut-and-tag\"\n >15-1017</a\n >). The gel was imaged and Cy5 emission was observed at 670\n nm (right), stained with ethidium bromide, and imaged again\n at 605 nm. The observed shift indicates successful loading\n of the Tn5, and fluorescent imaging confirms the presence of\n the dye.\n </span>\n </p>\n </div>\n <div class=\"image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg\n class=\"image-picker__svg-left\"\n width=\"24\"\n height=\"24\"\n viewBox=\"0 0 24 24\">\n <path\n d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a\n href=\"/content/images/products/proteins/15-1026-size-distribution.jpeg\"\n target=\"blank\"\n class=\"image-picker__main-image-link\"\n ><img\n alt=\"15-1026-size-distribution\"\n src=\"/content/images/products/proteins/15-1026-size-distribution.jpeg\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\"\n >(Click to enlarge)</span\n ></a\n >\n <button class=\"image-picker__right-arrow\">\n <svg\n class=\"image-picker__svg-right\"\n width=\"24\"\n height=\"24\"\n viewBox=\"0 0 24 24\">\n <path\n d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\"\n ><strong\n >Figure 2: Size distribution of released chromatin</strong\n ><br />\n CUT&Tag was performed as described above. Recovered DNA was\n directly PCR amplified to produce sequence-ready libraries.\n Agilent TapeStation traces for libraries derived from\n negative control Rabbit IgG (top), H3K4me3 (middle), and\n H3K27me3 (bottom) are shown. Excised DNA is highly enriched\n for mononucleosomes (peak at ~300 bp reflects ~150 bp insert\n size).\n </span>\n </p>\n </div>\n <div class=\"image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg\n class=\"image-picker__svg-left\"\n width=\"24\"\n height=\"24\"\n viewBox=\"0 0 24 24\">\n <path\n d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a\n href=\"/content/images/products/proteins/15-1026-cut-tag-data.jpeg\"\n target=\"blank\"\n class=\"image-picker__main-image-link\">\n <img\n alt=\"15-1026-cut-tag-data\"\n src=\"/content/images/products/proteins/15-1026-cut-tag-data.jpeg\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\"\n >(Click to enlarge)</span\n >\n </a>\n <button class=\"image-picker__right-arrow\">\n <svg\n class=\"image-picker__svg-right\"\n width=\"24\"\n height=\"24\"\n viewBox=\"0 0 24 24\">\n <path\n d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\">\n <strong>Figure 3: CUT&Tag data </strong><br />\n CUT&Tag was performed as described above. Representative\n sequencing tracks obtained using CUTANA Fluorescent pAG-Tn5\n for CUT&Tag show a 337 kb view of the TRMT2A gene. CUTANA\n pAG-Tn5 produced clear peaks with genomic distribution\n consistent with the known biological functions of H3K4me3\n and H3K27me3 as well as minimal background in the IgG\n negative control.\n </span>\n </p>\n </div>\n <div class=\"image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg\n class=\"image-picker__svg-left\"\n width=\"24\"\n height=\"24\"\n viewBox=\"0 0 24 24\">\n <path\n d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a\n href=\"/content/images/products/proteins/15-1026-protein-gel-data.jpeg\"\n target=\"blank\"\n class=\"image-picker__main-image-link\">\n <img\n alt=\"15-1026-protein-gel-data\"\n src=\"/content/images/products/proteins/15-1026-protein-gel-data.jpeg\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\"\n >(Click to enlarge)</span\n >\n </a>\n <button class=\"image-picker__right-arrow\">\n <svg\n class=\"image-picker__svg-right\"\n width=\"24\"\n height=\"24\"\n viewBox=\"0 0 24 24\">\n <path\n d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\">\n <strong>Figure 4: Protein gel data </strong><br />\n CUTANA pAG-Tn5 (1 µg) was resolved via SDS-PAGE and stained\n with Coomassie blue. The migration and molecular weight of\n the protein standards are indicated. Uncharged pAG-Tn5\n monomer is 78.5 kDa, however once charged with DNA, Tn5\n dimerizes to a final complex weight of 192 kDa.\n </span>\n </p>\n </div>\n <div class=\"image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg\n class=\"image-picker__svg-left\"\n width=\"24\"\n height=\"24\"\n viewBox=\"0 0 24 24\">\n <path\n d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a\n href=\"/content/images/products/methods/cut-and-tag-methods.png\"\n target=\"blank\"\n class=\"image-picker__main-image-link\">\n <img\n alt=\"cut-and-tag-methods\"\n src=\"/content/images/products/methods/cut-and-tag-methods.png\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\"\n >(Click to enlarge)</span\n >\n </a>\n <button class=\"image-picker__right-arrow\">\n <svg\n class=\"image-picker__svg-right\"\n width=\"24\"\n height=\"24\"\n viewBox=\"0 0 24 24\">\n <path\n d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\">\n <strong>CUT&Tag methods</strong><br />\n CUT&Tag was performed on 100k K562 cells with 0.5 µg of\n either IgG (EpiCypher\n <a\n href=\"/products/nucleosomes/snap-cutana-spike-in-controls/cutana-rabbit-igg-cut-run-negative-control-antibody\"\n >13-0042</a\n >), H3K4me3 (EpiCypher\n <a\n href=\"/products/antibodies/snap-chip-certified-antibodies/histone-h3k4me3-antibody-snap-chip-certified-cutana-cut-run-compatible\"\n >13-0041</a\n >), or H3K27me3 (ThermoFisher\n <a\n href=\"https://www.thermofisher.com/antibody/product/H3K27me3-Antibody-clone-G-299-10-Monoclonal/MA5-11198\"\n >MA5-11198</a\n >) antibodies using CUTANA™ Fluorescent pAG-Tn5 (50 nM\n final; see Technical Datasheet for lot-specific\n recommendation) following the EpiCypher Direct-to-PCR\n CUT&Tag <a href=\"/protocols\">protocol</a>. Libraries were\n run on an Illumina NextSeq2000 with paired-end sequencing\n (2x50 bp). Sample sequencing depth was 0.5 million reads\n (IgG), 2.5 million reads (H3K4me3), and 6.3 million reads\n (H3K27me3). Data were aligned to the hg19 genome using\n Bowtie2. Data were filtered to remove duplicates,\n multi-aligned reads, and ENCODE DAC Exclusion List regions.\n </span>\n </p>\n </div>\n </div>\n <aside class=\"image-picker__right\">\n <div class=\"image-picker__gallery\">\n <img\n alt=\"15-1026-protein-charging-data\"\n src=\"/content/images/products/proteins/15-1026-charging-data.jpeg\"\n width=\"200\"\n class=\"image-picker__side-image image-picker__side-image_active\"\n role=\"button\" />\n <img\n alt=\"15-1026-size-distribution\"\n src=\"/content/images/products/proteins/15-1026-size-distribution.jpeg\"\n class=\"image-picker__side-image\"\n role=\"button\" />\n <img\n alt=\"15-1026-cut-tag-data\"\n src=\"/content/images/products/proteins/15-1026-cut-tag-data.jpeg\"\n class=\"image-picker__side-image\"\n role=\"button\" />\n <img\n alt=\"15-1026-protein-gel-data\"\n src=\"/content/images/products/proteins/15-1026-protein-gel-data.jpeg\"\n width=\"200\"\n class=\"image-picker__side-image image-picker__side-image_active\"\n role=\"button\" />\n <img\n alt=\"cut-and-tag-methods\"\n src=\"/content/images/products/methods/cut-and-tag-methods.png\"\n width=\"200\"\n class=\"image-picker__side-image image-picker__side-image_active\"\n role=\"button\" />\n </div>\n </aside>\n </section>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Recommended Accessory Products</h3>\n <div\n class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <table class=\"epicypher-table\">\n <tr>\n <th>Item</th>\n <th>Cat. No.</th>\n </tr>\n\n <tr>\n <td>Anti-Rabbit Secondary Antibody for CUTANA™ CUT&Tag</td>\n <td>\n <a\n href=\"/products/epigenetics-reagents-and-assays/cutana-cut-tag-assays/anti-rabbit-secondary-antibody-for-cutana-cut-tag-workflows\"\n >13-0047</a\n >\n </td>\n </tr>\n\n <tr>\n <td>Anti-Mouse Secondary Antibody for CUTANA™ CUT&Tag</td>\n <td>\n <a\n href=\"/products/epigenetics-reagents-and-assays/cutanac-cut-tag-assays/anti-mouse-secondary-antibody-for-cutana-cut-tag-workflows\"\n >13-0048</a\n >\n </td>\n </tr>\n\n <tr>\n <td>CUTANA™ Non-Hot Start 2X PCR Master Mix for CUT&Tag</td>\n <td>\n <a\n href=\"/products/epigenetics-reagents-and-assays/cutana-chic-cut-tag-assays/cutana-high-fidelity-2x-pcr-master-mix\"\n >15-1018</a\n >\n </td>\n </tr>\n\n <tr>\n <td>H3K4me3 Antibody, SNAP-Certified™ for CUT&RUN and CUT&Tag</td>\n <td>\n <a\n href=\"/products/antibodies/h3k4me3-antibody-snap-certified-for-cut-run-and-cut-tag\"\n >13-0060</a\n >\n </td>\n </tr>\n\n <tr>\n <td>CUTANA™ Rabbit IgG CUT&RUN Negative Control Antibody</td>\n <td>\n <a\n href=\"/products/nucleosomes/snap-cutana-spike-in-controls/cutana-rabbit-igg-cut-run-negative-control-antibody\"\n >13-0042</a\n >\n </td>\n </tr>\n </table>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Technical Information</h3>\n <div\n class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <div class=\"product-tech-info\">\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Storage</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n Stable for one year at -20°C from date of receipt. The protein\n is not subject to freeze/thaw under these conditions.\n </div>\n </div>\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Formulation</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n 50 mM HEPES-KOH pH 7.2, 100 mM NaCl, 0.1 mM EDTA, 1 mM DTT, 0.1%\n Triton X-100, 50% glycerol\n </div>\n </div>\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Adapters</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n <p style=\"margin-bottom: 0\">\n Tn5ME-A: 5’-[Cy5]TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG-3’\n </p>\n <p style=\"margin-bottom: 0\">\n Tn5ME-B: 5’-[Cy5]GATTCGTGGGCTCGGAGATGTGTATAAGAGACAG-3’\n </p>\n <p style=\"margin-bottom: 0\">\n Tn5ME-rev: 5’-[phos]CTGTCTCTTATACACATCT-3’\n </p>\n </div>\n </div>\n </div>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Application Notes</h3>\n <div\n class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <p>\n A 20X dilution of this reagent is sufficient for a standard CUT&Tag\n reaction following the EpiCypher CUTANA™ Direct-to-PCR CUT&Tag\n <a href=\"/protocols\">protocol</a>, indicating that addition of the\n fluorescent dye does not prevent Tn5-mediated tagmentation of\n chromatin (see Figures 2 & 3). However, application-specific\n optimization will be necessary for use in microscopy. Due to the\n novelty of this reagent, EpiCypher will not engage in application\n tech support.\n </p>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">References</h3>\n <div\n class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <b>Background References:</b><br />\n [1] Kaya-Okur et al. <em>Nat. Commun.</em> (2019). PMID:\n <a\n href=\"https://pubmed.ncbi.nlm.nih.gov/31036827/\"\n title=\"CUT&Tag for efficient epigenomic profiling of small samples and single cells\"\n target=\"new\"\n >31036827</a\n ><br />\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Documents & Resources</h3>\n <div\n class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <div class=\"product-documents\">\n <a\n href=\"/content/documents/tds/15-1026.pdf\"\n target=\"blank\"\n class=\"product-documents__link\">\n <svg\n version=\"1.1\"\n id=\"Layer_1\"\n xmlns=\"http://www.w3.org/2000/svg\"\n xmlns:xlink=\"http://www.w3.org/1999/xlink\"\n x=\"0px\"\n y=\"0px\"\n viewBox=\"0 0 228 240\"\n style=\"enable-background: new 0 0 228 240\"\n xml:space=\"preserve\"\n class=\"product-documents__icon\">\n <g>\n <path\n class=\"product-documents__svg-pdf\"\n d=\"M191.92,68.77l-47.69-47.69c-1.33-1.33-3.12-2.08-5.01-2.08H45.09C41.17,19,38,22.17,38,26.09v184.36\n c0,3.92,3.17,7.09,7.09,7.09h141.82c3.92,0,7.09-3.17,7.09-7.09V73.8C194,71.92,193.25,70.1,191.92,68.77z M177.65,77.06h-41.7\n v-41.7L177.65,77.06z M178.05,201.59H53.95V34.95h66.92v47.86c0,5.14,4.17,9.31,9.31,9.31h47.86V201.59z\" />\n </g>\n <rect\n x=\"20\"\n y=\"112\"\n class=\"product-documents__svg-background\"\n width=\"146\"\n height=\"76\" />\n <g>\n <path\n class=\"product-documents__svg-pdf\"\n d=\"M23.83,125.68h22.36c5.29,0,9.41,1.33,12.35,4c2.94,2.67,4.42,6.39,4.42,11.18c0,4.78-1.47,8.51-4.42,11.18\n c-2.94,2.67-7.06,4-12.35,4H34.59v18.29H23.83V125.68z M44.81,147.9c5.38,0,8.07-2.32,8.07-6.97c0-2.39-0.67-4.16-2-5.31\n c-1.33-1.15-3.36-1.73-6.07-1.73H34.59v14.01H44.81z\" />\n <path\n class=\"product-documents__svg-pdf\"\n d=\"M69.92,125.68h18.91c5.29,0,9.84,0.97,13.66,2.9c3.82,1.93,6.74,4.72,8.76,8.35\n c2.02,3.63,3.04,7.98,3.04,13.04c0,5.06-1,9.42-3,13.08c-2,3.66-4.91,6.45-8.73,8.38c-3.82,1.93-8.4,2.9-13.73,2.9H69.92V125.68z\n M88.07,165.63c10.35,0,15.52-5.22,15.52-15.66c0-10.4-5.17-15.59-15.52-15.59h-7.38v31.26H88.07z\" />\n <path\n class=\"product-documents__svg-pdf\"\n d=\"M122.57,125.68h32.84v8.49h-22.22v11.18h20.84v8.49h-20.84v20.49h-10.63V125.68z\" />\n </g>\n </svg>\n <span class=\"product-documents__info\">Technical Datasheet</span>\n </a>\n </div>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Intellectual Property</h3>\n <div\n class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <p>\n US Pat. No. 10,689,643, 11,306,307, EU Pat. No. 3,688,157, 2,999,784\n and related patents and pending applications\n </p>\n </div>\n </div>\n </div>\n </div>\n</div>\n\n<style>\n .form-field-title .required-text {\n display: none !important;\n }\n\n .form-label {\n padding-left: 2rem;\n }\n\n .form-label-text {\n margin: 0;\n margin-left: 0 !important;\n }\n\n /* //////////// */\n\n /* BIOZ */\n td table {\n margin: 0;\n padding: 6px !important;\n }\n\n span.bioz-w-add-underline {\n font-size: 15px;\n }\n\n /* /////////// */\n\n .form-field-title .required-text {\n display: none !important;\n }\n .form-label {\n padding-left: 2rem;\n }\n .form-label-text {\n margin: 0;\n margin-left: 0 !important;\n }\n\n .bioz-w-header td {\n padding: 0;\n }\n\n .epicypher-table table {\n margin-right: auto;\n margin-left: auto;\n width: 100%;\n border-top: 1px solid lightgray;\n }\n\n .epicypher-table th {\n color: #3b3a48;\n }\n .epicypher-table td {\n color: #3b3a48;\n font-size: 0.9rem;\n }\n .epicypher-table td a {\n text-decoration: underline;\n }\n .epicypher-table td,\n th {\n border: 0.5px solid lightgray;\n text-align: left;\n padding: 8px;\n /* white-space: nowrap; */\n }\n .epicypher-table tr:nth-child(even) {\n background-color: #f9f9f9;\n }\n .epicypher-table td:first-child {\n border-left: 1px solid lightgray;\n }\n /* tr:hover {\n background-color: #ffff99;\n } */\n\n /* Tablet */\n\n @media only screen and (max-width: 760px),\n (min-device-width: 760px) and (max-device-width: 1024px) {\n /* Force table to not be like tables anymore */\n .epicypher-table table,\n thead,\n tbody,\n th,\n td,\n tr {\n display: block;\n }\n .epicypher-table th {\n display: none !important;\n }\n\n /* BIOZ */\n td.bioz-w-tooltipx::before {\n display: none;\n }\n\n /* .epicypher-table table,\n thead,\n tbody,\n th,\n td,\n tr {\n display: inline-table !important;\n } */\n\n /* Hide table headers (but not display: none;, for accessibility) */\n .epicypher-table thead tr {\n position: absolute;\n top: -9999px;\n left: -9999px;\n }\n\n .epicypher-table tr {\n border: 1px solid #ccc;\n }\n\n .epicypher-table td {\n /* Behave like a \"row\" */\n /* border: none; */\n border-bottom: 1px solid #eee;\n position: relative;\n padding-left: 30%;\n white-space: inherit;\n }\n\n .epicypher-table td:before {\n /* Now like a table header */\n position: absolute;\n /* Top/left values mimic padding */\n top: 6px;\n left: 6px;\n width: 45%;\n padding-right: 10px;\n white-space: nowrap;\n font-weight: 700;\n }\n\n /*\n Label the data\n */\n .epicypher-table td:nth-of-type(1):before {\n content: 'Item';\n }\n td:nth-of-type(2):before {\n content: 'Cat. No.';\n }\n }\n\n /* @media screen and (max-width: 767px) {\n .custom-title-description {\n font-size: 19px !important;\n }\n .section-title {\n font-size: 19px !important;\n padding-left: 1rem;\n }\n } */\n\n @media screen and (min-width: 768px) {\n .products-featured .container,\n .products-featured .product-tabs,\n .products-related .container,\n .products-related .product-tabs {\n padding-left: 30px !important;\n padding-right: 50px;\n max-width: 1170px !important;\n }\n }\n\n @media screen and (max-width: 767px) {\n .custom-title-description {\n font-size: 19px !important;\n }\n\n .section-title {\n font-size: 19px !important;\n padding-left: 1rem;\n }\n }\n</style>\n","tags":[],"warranty":"","price":{"without_tax":{"formatted":"$745.00","value":745,"currency":"USD"},"tax_label":"Sales Tax"},"detail_messages":"","availability":"","page_title":"CUTANA™ pAG-Tn5 for CUT&Tag, Fluorescent","cart_url":"https://www.epicypher.com/cart.php","max_purchase_quantity":0,"mpn":null,"upc":null,"options":[],"related_products":[{"id":764,"sku":null,"name":"CUTANA™ pAG-Tn5 for CUT&Tag","url":"https://www.epicypher.com/products/epigenetics-kits-and-reagents/cutana-pag-tn5-for-cut-and-tag","availability":"","rating":null,"brand":{"name":null},"category":["Epigenetics Kits and Reagents","Epigenetics Kits and Reagents/CUTANA™ CUT&Tag Assays"],"summary":"\n \n \n Type: Transposase\n \n \n Mol Wgt: 191 kDa\n \n \n \n ","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/764/750/2020_CUTTag_icon_RGB_Andy_L__93238.1592491196.png?c=2","alt":"CUTANA™ pAG-Tn5 for CUT&Tag"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/764/750/2020_CUTTag_icon_RGB_Andy_L__93238.1592491196.png?c=2","alt":"CUTANA™ pAG-Tn5 for CUT&Tag"}],"date_added":"17th Jun 2020","pre_order":false,"show_cart_action":true,"has_options":true,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":[{"id":1005,"name":"Internal Comment","value":"extra boxes in bottom shelf of Venom"}],"num_reviews":null,"weight":{"formatted":"0.01 LBS","value":0.01},"demo":false,"price":{"without_tax":{"currency":"USD","formatted":"$795.00","value":795},"price_range":{"min":{"without_tax":{"currency":"USD","formatted":"$795.00","value":795},"tax_label":"Sales Tax"},"max":{"without_tax":{"currency":"USD","formatted":"$2,995.00","value":2995},"tax_label":"Sales Tax"}},"tax_label":"Sales Tax"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=764"},{"id":766,"sku":"15-1025","name":"CUTANA™ Uncharged pAG-Tn5 for CUT&Tag","url":"https://www.epicypher.com/products/epigenetics-kits-and-reagents/cutana-cut-tag-assays/cutana-uncharged-pag-tn5-for-cut-tag","availability":"","rating":null,"brand":{"name":null},"category":["Epigenetics Kits and Reagents","Epigenetics Kits and Reagents/CUTANA™ CUT&Tag Assays","Other/IDEA Toolbox"],"summary":"\n \n \n Type: Transposase\n \n \n Mol Wgt: 157 kDa (dimer)\n \n \n \n ","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/766/1065/toolbox-icon__65890.1661116229__53021.1661977262.png?c=2","alt":"CUTANA™ Uncharged pAG-Tn5 for CUT&Tag"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/766/1065/toolbox-icon__65890.1661116229__53021.1661977262.png?c=2","alt":"CUTANA™ Uncharged pAG-Tn5 for CUT&Tag"}],"date_added":"14th Jul 2020","pre_order":false,"show_cart_action":true,"has_options":false,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":[{"id":1206,"name":"Pack Size","value":"20 µL"}],"num_reviews":null,"weight":{"formatted":"0.01 LBS","value":0.01},"demo":false,"add_to_cart_url":"https://www.epicypher.com/cart.php?action=add&product_id=766","price":{"without_tax":{"currency":"USD","formatted":"$645.00","value":645},"tax_label":"Sales Tax"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=766"},{"id":1160,"sku":"15-1028","name":"CUTANA™ Uncharged 6xHis-pAG-Tn5 for CUT&Tag","url":"https://www.epicypher.com/products/epigenetics-kits-and-reagents/cutana-cut-tag-assays/cutana-uncharged-6xhis-pag-tn5-for-cut-tag","availability":"","rating":null,"brand":{"name":null},"category":["Epigenetics Kits and Reagents","Epigenetics Kits and Reagents/CUTANA™ CUT&Tag Assays","Other/IDEA Toolbox"],"summary":"\n \n \n Type: Transposase\n \n \n Mol Wgt: 161,401.8 Da (dimer)\n \n \n \n ","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/1160/1233/toolbox-icon__65890.1661116229__13690.1718896281.png?c=2","alt":"CUTANA™ Uncharged 6xHis-pAG-Tn5 for CUT&Tag"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/1160/1233/toolbox-icon__65890.1661116229__13690.1718896281.png?c=2","alt":"CUTANA™ Uncharged 6xHis-pAG-Tn5 for CUT&Tag"}],"date_added":"20th Jun 2024","pre_order":false,"show_cart_action":true,"has_options":false,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":[{"id":1326,"name":"Pack Size","value":"75 µL"}],"num_reviews":null,"weight":{"formatted":"0.01 LBS","value":0.01},"demo":false,"add_to_cart_url":"https://www.epicypher.com/cart.php?action=add&product_id=1160","price":{"without_tax":{"currency":"USD","formatted":"$645.00","value":645},"tax_label":"Sales Tax"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=1160"},{"id":986,"sku":null,"name":"CUTANA™ CUT&Tag Kit","url":"https://www.epicypher.com/products/epigenetics-kits-and-reagents/cutana-cut-and-tag-kit","availability":"","rating":null,"brand":{"name":null},"category":["Antibodies/CUTANA™ CUT&Tag Antibodies","Epigenetics Kits and Reagents/CUTANA™ CUT&Tag Assays"],"summary":"\n \n \n \n \n ","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/986/1213/CTK20__46720.1713641110.png?c=2","alt":"CUTANA™ CUT&Tag Kit"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/986/1213/CTK20__46720.1713641110.png?c=2","alt":"CUTANA™ CUT&Tag Kit"}],"date_added":"8th Mar 2023","pre_order":false,"show_cart_action":true,"has_options":true,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":[{"id":1219,"name":"Pack Size","value":"48 Reactions"}],"num_reviews":null,"weight":{"formatted":"0.01 LBS","value":0.01},"demo":false,"price":{"without_tax":{"currency":"USD","formatted":"$2,695.00","value":2695},"tax_label":"Sales Tax"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=986"},{"id":694,"sku":null,"name":"CUTANA™ pAG-MNase for ChIC/CUT&RUN Workflows","url":"https://www.epicypher.com/products/epigenetics-reagents-and-assays/cutana-pag-mnase-for-chic-cut-and-run-workflows","availability":"","rating":null,"brand":{"name":null},"category":["Epigenetics Kits and Reagents","Epigenetics Kits and Reagents/CUTANA™ ChIC / CUT&RUN Assays"],"summary":"\n \n \n Type: Nuclease\n \n \n Mol Wgt: 43.7 kDa\n \n \n \n ","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/694/689/Screen_Shot_2020-02-12_at_11.01.55_AM__17144.1581530752.png?c=2","alt":"CUTANA™ pAG-MNase for ChIC/CUT&RUN Workflows"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/694/689/Screen_Shot_2020-02-12_at_11.01.55_AM__17144.1581530752.png?c=2","alt":"CUTANA™ pAG-MNase for ChIC/CUT&RUN Workflows"}],"date_added":"12th Aug 2019","pre_order":false,"show_cart_action":true,"has_options":true,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":[{"id":1174,"name":"Internal Comment","value":"Excess in bottom of Venom"},{"id":1175,"name":"Internal Comment","value":"Bulk in Psylocke"}],"num_reviews":null,"weight":{"formatted":"0.01 LBS","value":0.01},"demo":false,"price":{"without_tax":{"currency":"USD","formatted":"$335.00","value":335},"price_range":{"min":{"without_tax":{"currency":"USD","formatted":"$335.00","value":335},"tax_label":"Sales Tax"},"max":{"without_tax":{"currency":"USD","formatted":"$1,295.00","value":1295},"tax_label":"Sales Tax"}},"tax_label":"Sales Tax"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=694"}],"shipping_messages":[],"rating":0,"meta_keywords":"CUT&Tag, pAG-Tn5, Tn5, transposase, tagmentation, cutandtag, chromatin mapping, microscopy, chromatin structure, epigenomic mapping, ChIP-seq, Cleavage Under Targets and Tagmentation","show_quantity_input":1,"title":"CUTANA™ Fluorescent pAG-Tn5 for CUT&Tag","gift_wrapping_available":false,"min_purchase_quantity":0,"customizations":[],"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/934/1048/toolbox-icon__95574.1661116173.png?c=2","alt":"CUTANA™ Fluorescent pAG-Tn5 for CUT&Tag"}]} Pack Size: 125 µL
- Type: Transposase
- Mol Wgt: 192 kDa
- Host: E. coli
- Epitope Tag: None
Description
Products in EpiCypher’s IDEA Toolbox (Innovation and Discovery of Epigenetic Applications) offer access to reagents without known or fully defined uses, enabling researchers to explore cutting-edge applications. Due to their novelty and unexplored potential, EpiCypher will engage in limited technical support.
CUTANA Fluorescent pAG-Tn5 is a fusion of Proteins A and G to E. coli transposase (Tn5), the key enzyme for CUT&Tag [1]. The Mosaic A and Mosaic B adapters loaded in this enzyme bear a 5’ Cy5 fluorescent dye to enable multiomic visualization and mapping of chromatin features. This product is highly purified to remove contaminating E. coli DNA. For an enzyme charged with non-fluorescent adapters that can be used in standard CUT&Tag, see EpiCypher 15-1017.
Validation Data
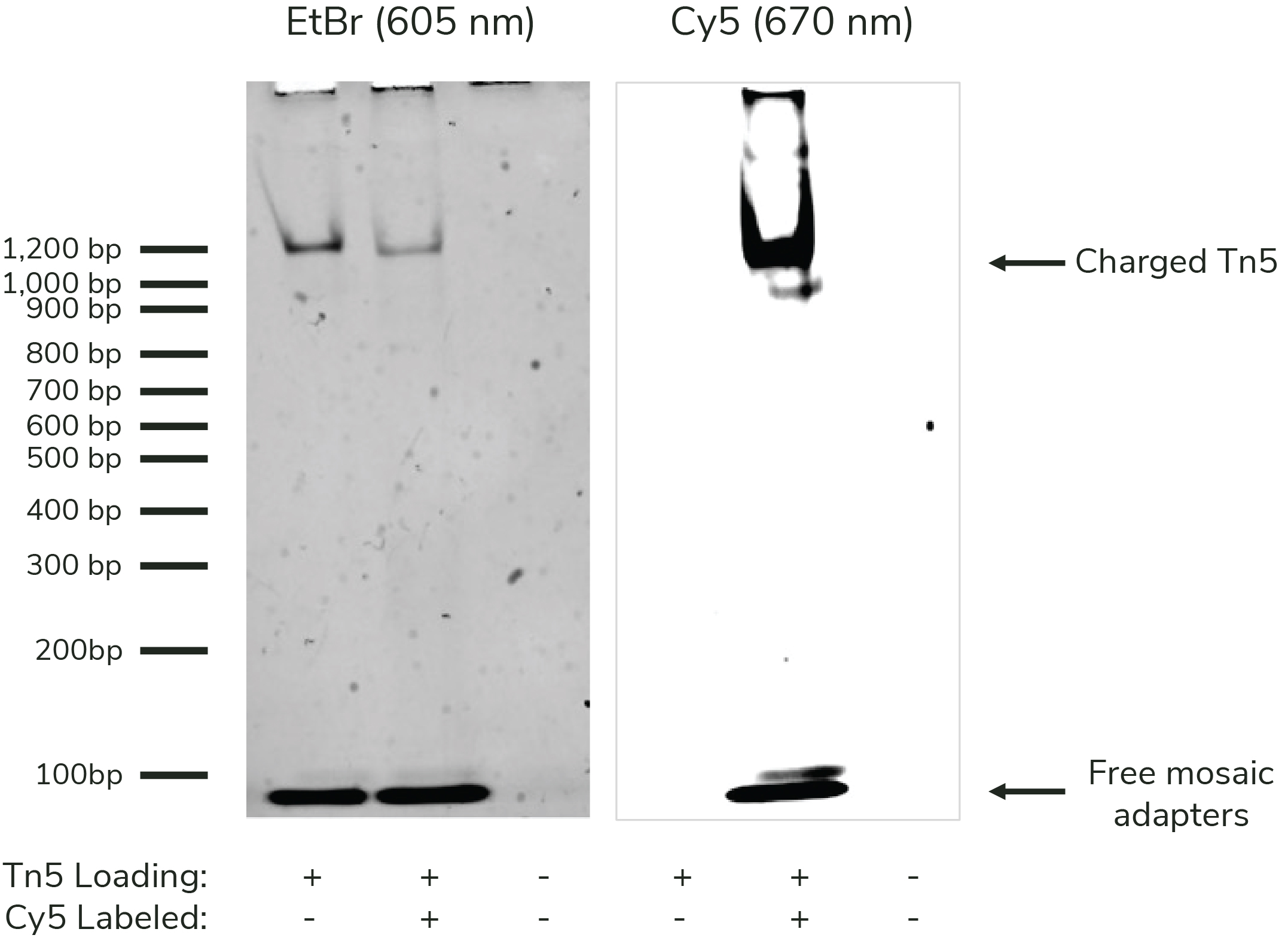
Figure 1: DNA charging data
CUTANA Fluorescent pAG-Tn5 for CUT&Tag (0.5 µg) was resolved
via Native PAGE alongside uncharged pAG-Tn5 (EpiCypher
15-1025) and charged pAG-Tn5 (EpiCypher
15-1017). The gel was imaged and Cy5 emission was observed at 670
nm (right), stained with ethidium bromide, and imaged again
at 605 nm. The observed shift indicates successful loading
of the Tn5, and fluorescent imaging confirms the presence of
the dye.
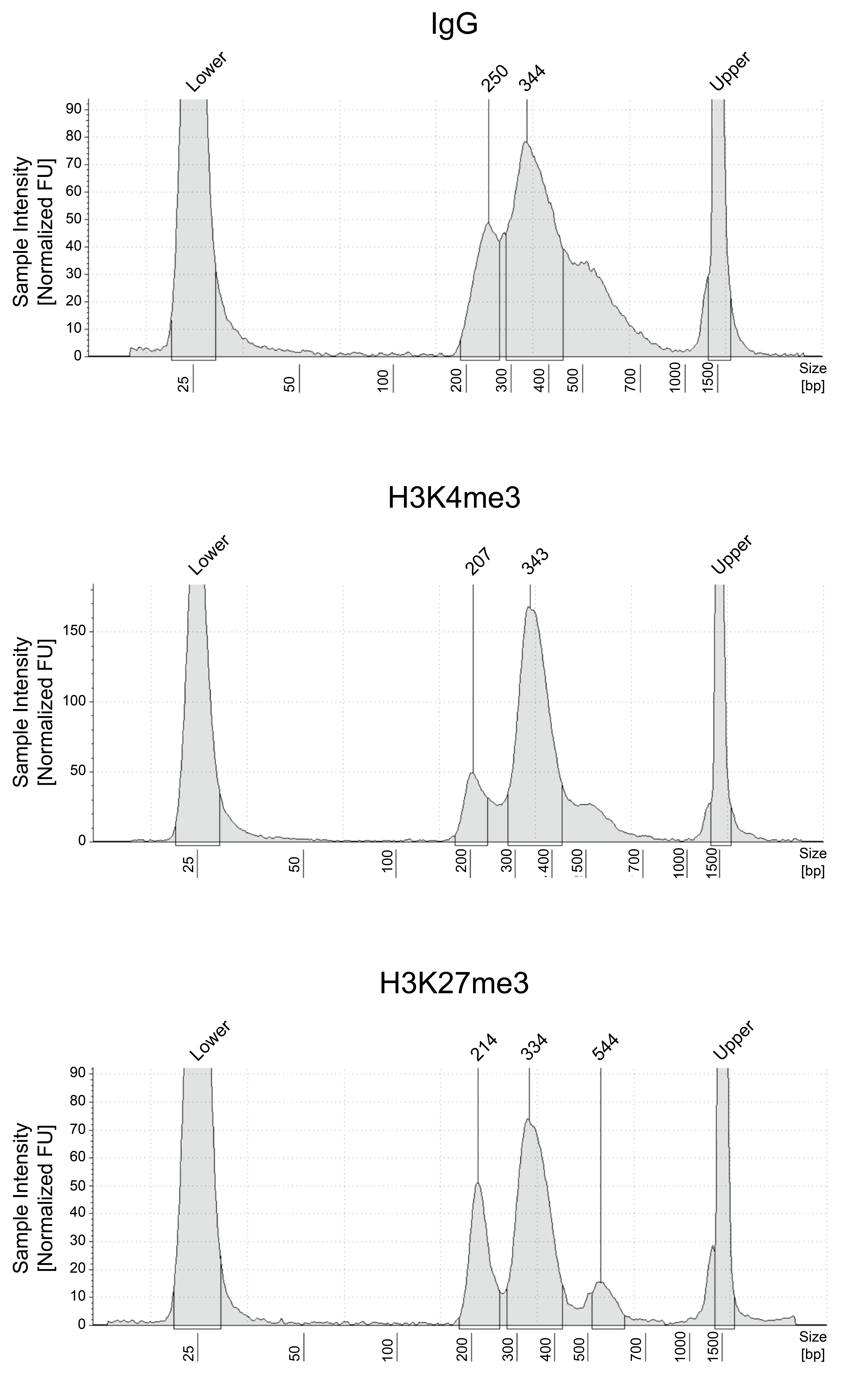
Figure 2: Size distribution of released chromatin
CUT&Tag was performed as described above. Recovered DNA was
directly PCR amplified to produce sequence-ready libraries.
Agilent TapeStation traces for libraries derived from
negative control Rabbit IgG (top), H3K4me3 (middle), and
H3K27me3 (bottom) are shown. Excised DNA is highly enriched
for mononucleosomes (peak at ~300 bp reflects ~150 bp insert
size).
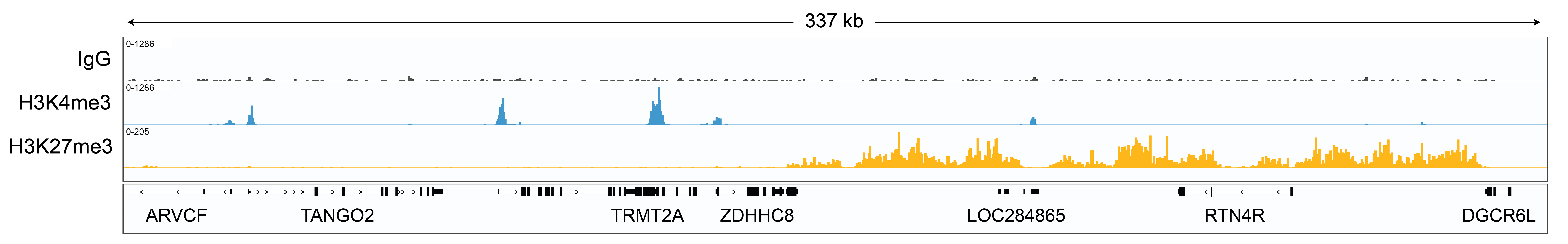
Figure 3: CUT&Tag data
CUT&Tag was performed as described above. Representative
sequencing tracks obtained using CUTANA Fluorescent pAG-Tn5
for CUT&Tag show a 337 kb view of the TRMT2A gene. CUTANA
pAG-Tn5 produced clear peaks with genomic distribution
consistent with the known biological functions of H3K4me3
and H3K27me3 as well as minimal background in the IgG
negative control.
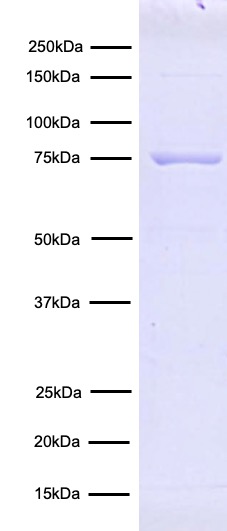
Figure 4: Protein gel data
CUTANA pAG-Tn5 (1 µg) was resolved via SDS-PAGE and stained
with Coomassie blue. The migration and molecular weight of
the protein standards are indicated. Uncharged pAG-Tn5
monomer is 78.5 kDa, however once charged with DNA, Tn5
dimerizes to a final complex weight of 192 kDa.
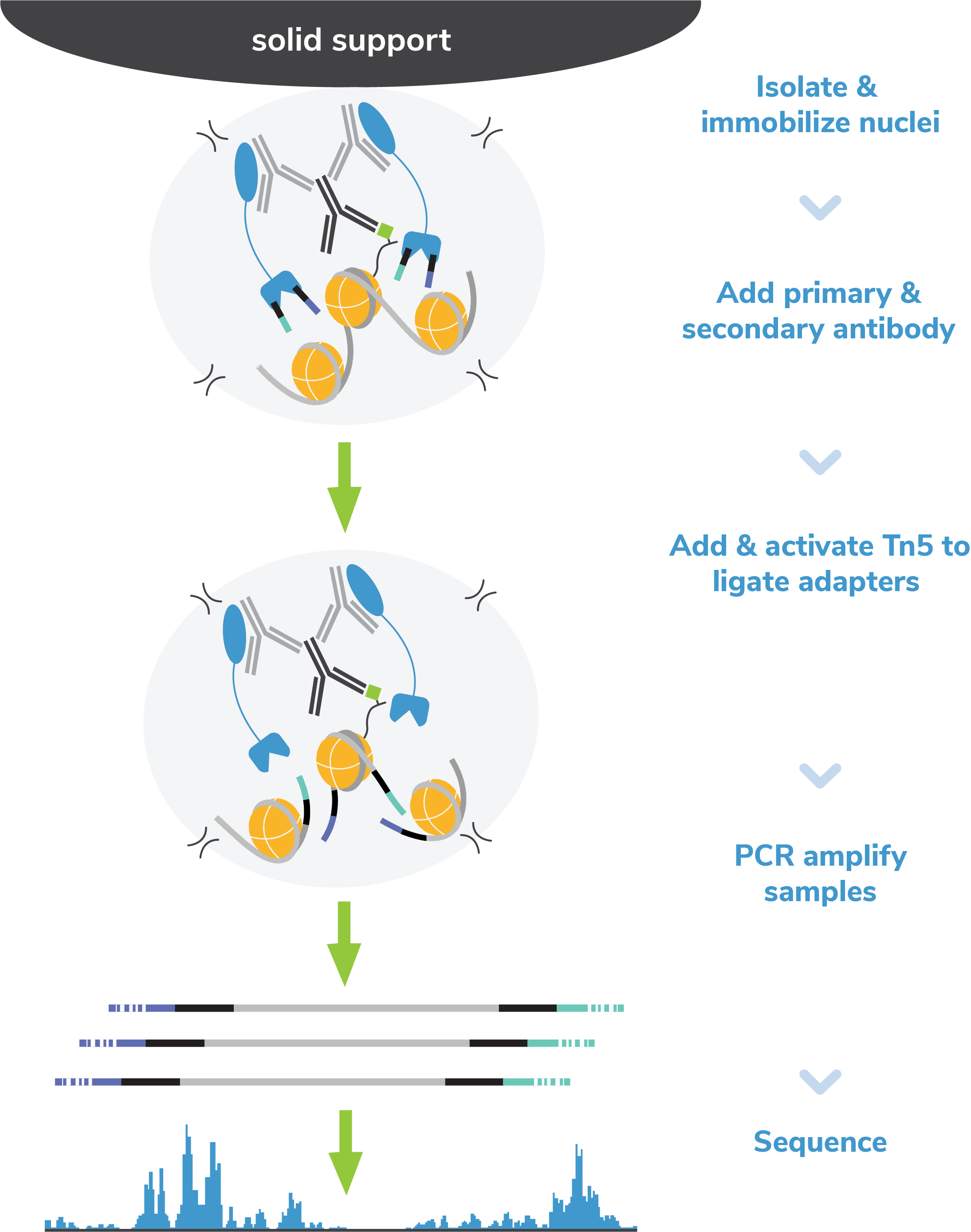
CUT&Tag methods
CUT&Tag was performed on 100k K562 cells with 0.5 µg of
either IgG (EpiCypher
13-0042), H3K4me3 (EpiCypher
13-0041), or H3K27me3 (ThermoFisher
MA5-11198) antibodies using CUTANA™ Fluorescent pAG-Tn5 (50 nM
final; see Technical Datasheet for lot-specific
recommendation) following the EpiCypher Direct-to-PCR
CUT&Tag protocol. Libraries were
run on an Illumina NextSeq2000 with paired-end sequencing
(2x50 bp). Sample sequencing depth was 0.5 million reads
(IgG), 2.5 million reads (H3K4me3), and 6.3 million reads
(H3K27me3). Data were aligned to the hg19 genome using
Bowtie2. Data were filtered to remove duplicates,
multi-aligned reads, and ENCODE DAC Exclusion List regions.
Recommended Accessory Products
| Item | Cat. No. |
|---|---|
| Anti-Rabbit Secondary Antibody for CUTANA™ CUT&Tag | 13-0047 |
| Anti-Mouse Secondary Antibody for CUTANA™ CUT&Tag | 13-0048 |
| CUTANA™ Non-Hot Start 2X PCR Master Mix for CUT&Tag | 15-1018 |
| H3K4me3 Antibody, SNAP-Certified™ for CUT&RUN and CUT&Tag | 13-0060 |
| CUTANA™ Rabbit IgG CUT&RUN Negative Control Antibody | 13-0042 |
Technical Information
Tn5ME-A: 5’-[Cy5]TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG-3’
Tn5ME-B: 5’-[Cy5]GATTCGTGGGCTCGGAGATGTGTATAAGAGACAG-3’
Tn5ME-rev: 5’-[phos]CTGTCTCTTATACACATCT-3’
Application Notes
A 20X dilution of this reagent is sufficient for a standard CUT&Tag reaction following the EpiCypher CUTANA™ Direct-to-PCR CUT&Tag protocol, indicating that addition of the fluorescent dye does not prevent Tn5-mediated tagmentation of chromatin (see Figures 2 & 3). However, application-specific optimization will be necessary for use in microscopy. Due to the novelty of this reagent, EpiCypher will not engage in application tech support.
References
Documents & Resources
Intellectual Property
US Pat. No. 10,689,643, 11,306,307, EU Pat. No. 3,688,157, 2,999,784 and related patents and pending applications