CUTANA™ IgG Negative Control Antibody for CUT&RUN and CUT&Tag
{"url":"https://www.epicypher.com/products/nucleosomes/snap-cutana-spike-in-controls/cutana-igg-negative-control-antibody-for-cut-run-and-cut-tag","add_this":[{"service":"facebook","annotation":""},{"service":"email","annotation":""},{"service":"print","annotation":""},{"service":"twitter","annotation":""},{"service":"linkedin","annotation":""}],"gtin":null,"id":758,"bulk_discount_rates":[],"can_purchase":true,"meta_description":"CUTANA™ Rabbit IgG CUT&RUN Negative Control Antibody for ChIC/CUT&RUN Assays","category":["Chromatin Mapping","Chromatin Mapping/CUTANA™ ChIC / CUT&RUN Assays","Chromatin Mapping/CUTANA™ CUT&Tag Assays","Antibodies/CUTANA™ CUT&RUN Compatible Antibodies","Nucleosomes/SNAP-CUTANA™ Spike-in Controls","Antibodies/CUTANA™ CUT&RUN Antibodies"],"AddThisServiceButtonMeta":"","main_image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/758/739/1579731877.1280.1280__61771.1592491196.png?c=2","alt":"CUTANA™ IgG Negative Control Antibody for CUT&RUN and CUT&Tag"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=758","shipping":{"calculated":true},"num_reviews":0,"weight":"0.01 LBS","custom_fields":[{"id":"1148","name":"Pack Size","value":"100 µg"},{"id":"1149","name":"Internal Comment","value":"bulk stocks from Thermo in cold room"}],"sku":"13-0042","description":"<div class=\"product-general-info\">\n <ul class=\"product-general-info__list-left\">\n <li class=\"product-general-info__list-item\">\n <strong>Type: </strong>Polyclonal\n </li>\n <li class=\"product-general-info__list-item\">\n <strong>Host: </strong>Rabbit\n </li>\n <li class=\"product-general-info__list-item\">\n <strong>Applications: </strong>CUT&RUN, CUT&Tag\n </li>\n </ul>\n <ul class=\"product-general-info__list-right\">\n <li class=\"product-general-info__list-item\">\n <strong>Reactivity: </strong>Negative Control\n </li>\n <li class=\"product-general-info__list-item\">\n <strong>Format: </strong>Affinity Purified IgG\n </li>\n <li class=\"product-general-info__list-item\">\n <strong>Target Size: </strong>N/A\n </li>\n </ul>\n <ul class=\"product-general-info__list-right\">\n <li class=\"product-general-info__list-item\">\n <a href=\"#bioz\">\n <div id=\"w-s-3835-13-0042\" style=\"\n width: max-content;\n height: 58px;\n position: relative;\n overflow-y: hidden;\n \"></div>\n <div id=\"bioz-w-pb-13-0042-div\">\n <a id=\"bioz-w-pb-13-0042\" style=\"font-size: 12px; color: transparent\" href=\"https://www.bioz.com/\"\n target=\"_blank\">\n <img src=\"https://cdn.bioz.com/assets/favicon.png\" style=\"\n width: 11px;\n height: 11px;\n vertical-align: baseline;\n padding-bottom: 0px;\n margin-left: 0px;\n margin-bottom: 0px;\n float: none;\n display: none;\n \" />\n </a>\n </div>\n </a>\n </li>\n </ul>\n</div>\n<div class=\"service_accordion product-droppdown\">\n <div class=\"container\">\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel current\">\n <h3 class=\"sub-title1\">Description</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description-specific\">\n <p>\n Cleavage Under Targets & Release Using Nuclease (CUT&RUN) and Cleavage Under Targets and Tagmentation\n (CUT&Tag) are next generation assays for genomic mapping of chromatin features to replace Chromatin\n Immunoprecipitation (ChIP-seq) [<a href=\"https://pubmed.ncbi.nlm.nih.gov/28079019/\" target=\"_blank\">Skene &\n Henikoff, 2017</a>]. Unlike in ChIP-seq, where baseline signal is established by sequencing the fragmented\n input chromatin, reactions performed with this rabbit IgG antibody are used to determine background signal\n arising from non-specific MNase digestion and Tn5 tagmentation. Pair with the H3K4me3 positive control\n antibody (EpiCypher <a\n href=\"https://www.epicypher.com/products/antibodies/h3k4me3-antibody-snap-certified-for-cut-run-and-cut-tag\">13-0060</a>)\n for a well-controlled experiment.\n </p>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel current\">\n <h3 class=\"sub-title1\">Validation Data - CUT&RUN</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description-specific\">\n <section class=\"image-picker\">\n <div class=\"image-picker__left\">\n <div class=\"image-picker__main-content_active image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg class=\"image-picker__svg-left\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a href=\"/content/images/products/antibodies/13-0042-heat-map.jpg\"\n target=\"_new\" class=\"image-picker__main-image-link\"><img loading=\"lazy\" alt=\"13-0042 Heat Map\"\n src=\"/content/images/products/antibodies/13-0042-heat-map.jpg\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\">(Click to enlarge)</span></a>\n <button class=\"image-picker__right-arrow\">\n <svg class=\"image-picker__svg-right\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\"><strong>Figure 1: CUT&RUN genome wide\n enrichment</strong><br />\n CUT&RUN was performed as described in <strong>Figure 3</strong>. Heatmaps show H3K4me3 peaks\n relative to IgG antibody in aligned rows\n ranked by intensity (top to bottom) and colored such that red indicates high localized enrichment\n and blue denotes\n background signal. H3K4me3 antibody showed expected enrichment around the TSS. Despite some\n observable background in the\n IgG control, differential enrichment with the target antibody is as expected.\n </span>\n </p>\n </div>\n <div class=\"image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg class=\"image-picker__svg-left\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a href=\"/content/images/products/antibodies/13-0042-browser-tracks.jpg\"\n target=\"_new\" class=\"image-picker__main-image-link\"><img loading=\"lazy\" alt=\"13-0042 CUT&RUN Data\"\n src=\"/content/images/products/antibodies/13-0042-browser-tracks.jpg\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\">(Click to enlarge)</span></a>\n <button class=\"image-picker__right-arrow\">\n <svg class=\"image-picker__svg-right\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\"><strong>Figure 2: Rabbit IgG CUT&RUN tracks</strong><br />\n CUT&RUN was performed as described in <strong>Figure 3</strong>. Gene browser shots were generated\n using the Integrative Genomics Viewer (IGV, Broad Institute). Two gene loci show H3K4me3 peaks and\n minimal background in the IgG track, as expected.\n </span>\n </p>\n </div>\n <div class=\"image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg class=\"image-picker__svg-left\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a href=\"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/original/image-manager/13-0042-figure3.png?t=1701417003\"\n target=\"_new\" class=\"image-picker__main-image-link\"><img loading=\"lazy\"\n alt=\"13-0042-immunoprecipitation\"\n src=\"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/original/image-manager/13-0042-figure3.png?t=1701417003\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\">(Click to enlarge)</span></a>\n <button class=\"image-picker__right-arrow\">\n <svg class=\"image-picker__svg-right\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\"><strong>Figure 3: CUT&RUN methods</strong><br />\n CUT&RUN was performed on 500k native K562 cells with 0.5 µg of either IgG or H3K4me3 (EpiCypher\n <a href=\"/products/antibodies/h3k4me3-antibody-snap-certified-for-cut-run-and-cut-tag\" target=\"_blank\">13-0060</a>) antibodies in duplicate using the CUTANA™ ChIC/CUT&RUN Kit v4 (EpiCypher <a href=\"/products/epigenetics-reagents-and-assays/cutana-chic-cut-and-run-kit\" target=\"_blank\">14-1048</a>). Library\n preparation was performed with 5 ng of DNA (or the total amount recovered if less than 5 ng) using\n the CUTANA™ CUT&RUN Library Prep Kit (EpiCypher <a href=\"/products/epigenetics-reagents-and-assays/cutana-cut-and-run-library-prep-kit\" target=\"_blank\">14-1001/14-1002</a>). Both kit protocols were adapted\n for high throughput Tecan liquid handling. Sample sequencing depth was 7.8/6.8 million reads (IgG\n Rep 1/Rep 2) and 12.6/10.9 million reads (H3K4me3 Rep 1/Rep 2). Data were aligned to the hg19 genome\n using Bowtie2. Data were filtered to remove duplicates, multi-aligned reads, and ENCODE DAC\n Exclusion List regions.\n </span>\n </p>\n </div>\n </div>\n <aside class=\"image-picker__right\">\n <div class=\"image-picker__gallery\">\n <img loading=\"lazy\" alt=\"13-0042 Heat Map\"\n src=\"/content/images/products/antibodies/13-0042-heat-map.jpg\"\n width=\"200\" class=\"image-picker__side-image image-picker__side-image_active\" role=\"button\" />\n <img loading=\"lazy\" alt=\"13-0042 CUT&RUN Data\"\n src=\"/content/images/products/antibodies/13-0042-browser-tracks.jpg\"\n class=\"image-picker__side-image\" role=\"button\" />\n <img loading=\"lazy\" alt=\"13-0042-immunoprecipitation\"\n src=\"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/original/image-manager/13-0042-figure3.png?t=1701417003\"\n class=\"image-picker__side-image\" role=\"button\" />\n </div>\n </aside>\n </section>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Validation Data - CUT&Tag</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <section class=\"image-picker\">\n <div class=\"image-picker__left\">\n <div class=\"image-picker__main-content_active image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg class=\"image-picker__svg-left\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a href=\"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/original/image-manager/13-0042-figure4.jpeg?t=1701417006\"\n target=\"_new\" class=\"image-picker__main-image-link\"><img loading=\"lazy\" alt=\"13-0042 Heat Map\"\n src=\"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/original/image-manager/13-0042-figure4.jpeg?t=1701417006\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\">(Click to enlarge)</span></a>\n <button class=\"image-picker__right-arrow\">\n <svg class=\"image-picker__svg-right\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\"><strong>Figure 4: CUT&Tag genome wide\n enrichment</strong><br />\n CUT&Tag was performed as described in <strong>Figure 6</strong>. Heatmaps show H3K4me3 peaks\n relative to IgG antibody in aligned rows ranked by intensity (top to bottom) and colored such that\n red indicates high localized enrichment and blue denotes background signal. H3K4me3 antibody showed\n expected enrichment around the TSS, while the IgG antibody displayed minimal background, as\n expected.\n </span>\n </p>\n </div>\n <div class=\"image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg class=\"image-picker__svg-left\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a href=\"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/original/image-manager/13-0042-figure5.jpeg?t=1701417007\"\n target=\"_new\" class=\"image-picker__main-image-link\"><img loading=\"lazy\" alt=\"13-0042 CUT&RUN Data\"\n src=\"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/original/image-manager/13-0042-figure5.jpeg?t=1701417007\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\">(Click to enlarge)</span></a>\n <button class=\"image-picker__right-arrow\">\n <svg class=\"image-picker__svg-right\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\"><strong>Figure 5: Rabbit IgG CUT&Tag tracks</strong><br />\n CUT&Tag was performed as described in <strong>Figure 6</strong>. Gene browser shots were generated\n using the Integrative Genomics Viewer (IGV, Broad Institute). Two gene loci show H3K4me3 peaks and\n minimal background in the IgG track, as expected.\n </span>\n </p>\n </div>\n <div class=\"image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg class=\"image-picker__svg-left\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a href=\"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/original/image-manager/13-0042-figure6.png?t=1701417012\"\n target=\"_new\" class=\"image-picker__main-image-link\"><img loading=\"lazy\"\n alt=\"13-0042-immunoprecipitation\"\n src=\"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/original/image-manager/13-0042-figure6.png?t=1701417012\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\">(Click to enlarge)</span></a>\n <button class=\"image-picker__right-arrow\">\n <svg class=\"image-picker__svg-right\" width=\"24\" height=\"24\" viewBox=\"0 0 24 24\">\n <path d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\"><strong>Figure 6: CUT&Tag methods</strong><br />\n CUT&Tag was performed on 100k native K562 nuclei with 0.5 µg of either IgG or H3K4me3 (EpiCypher <a\n href=\"https://www.epicypher.com/products/antibodies/snap-chip-certified-antibodies/histone-h3k4me3-antibody-snap-chip-certified-cutana-cut-run-compatible\"\n target=\"_blank\">13-0041</a>) antibodies in singlicate using the CUTANA™ CUT&Tag Kit v1 (EpiCypher\n <a href=\"https://www.epicypher.com/products/epigenetics-kits-and-reagents/cutana-cut-and-tag-kit\"\n target=\"_blank\">14-1102/14-1103</a>). Libraries were run on an Illumina NextSeq2000 with\n paired-end sequencing (2x50bp). Sample sequencing depth was 0.9 million reads (IgG) and 2.9 million\n reads (H3K4me3). Data were aligned to the hg19 genome using Bowtie2. Data were filtered to remove\n duplicates, multi-aligned reads, and ENCODE DAC Exclusion List regions.\n </span>\n </p>\n </div>\n </div>\n <aside class=\"image-picker__right\">\n <div class=\"image-picker__gallery\">\n <img loading=\"lazy\" alt=\"13-0042 Heat Map\"\n src=\"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/original/image-manager/13-0042-figure4.jpeg?t=1701417006\"\n width=\"200\" class=\"image-picker__side-image image-picker__side-image_active\" role=\"button\" />\n <img loading=\"lazy\" alt=\"13-0042 CUT&RUN Data\"\n src=\"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/original/image-manager/13-0042-figure5.jpeg?t=1701417007\"\n class=\"image-picker__side-image\" role=\"button\" />\n <img loading=\"lazy\" alt=\"13-0042-immunoprecipitation\"\n src=\"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/original/image-manager/13-0042-figure6.png?t=1701417012\"\n class=\"image-picker__side-image\" role=\"button\" />\n </div>\n </aside>\n </section>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Technical Information</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <div class=\"product-tech-info\">\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Immunogen</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">None</div>\n </div>\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Storage</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n Stable for 1 year at 4°C from date of receipt\n </div>\n </div>\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Formulation</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n Affinity-purified antibody in PBS pH 7.6\n </div>\n </div>\n </div>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Recommended Dilution</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <div class=\"product-tech-info\">\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>CUT&RUN</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">0.5 µg per reaction</div>\n </div>\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>CUT&Tag</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n 0.5 µg per reaction\n </div>\n </div>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Documents & Resources</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <div class=\"product-documents\">\n <a href=\"/content/documents/tds/13-0042.pdf\" target=\"_new\" class=\"product-documents__link\">\n <svg version=\"1.1\" id=\"Layer_1\" xmlns=\"http://www.w3.org/2000/svg\"\n xmlns:xlink=\"http://www.w3.org/1999/xlink\" x=\"0px\" y=\"0px\" viewBox=\"0 0 228 240\"\n style=\"enable-background: new 0 0 228 240\" xml:space=\"preserve\" class=\"product-documents__icon\">\n <g>\n <path class=\"product-documents__svg-pdf\" d=\"M191.92,68.77l-47.69-47.69c-1.33-1.33-3.12-2.08-5.01-2.08H45.09C41.17,19,38,22.17,38,26.09v184.36\n\t\t\t c0,3.92,3.17,7.09,7.09,7.09h141.82c3.92,0,7.09-3.17,7.09-7.09V73.8C194,71.92,193.25,70.1,191.92,68.77z M177.65,77.06h-41.7\n\t\t\t v-41.7L177.65,77.06z M178.05,201.59H53.95V34.95h66.92v47.86c0,5.14,4.17,9.31,9.31,9.31h47.86V201.59z\" />\n </g>\n <rect x=\"20\" y=\"112\" class=\"product-documents__svg-background\" width=\"146\" height=\"76\" />\n <g>\n <path class=\"product-documents__svg-pdf\" d=\"M23.83,125.68h22.36c5.29,0,9.41,1.33,12.35,4c2.94,2.67,4.42,6.39,4.42,11.18c0,4.78-1.47,8.51-4.42,11.18\n\t\t\t c-2.94,2.67-7.06,4-12.35,4H34.59v18.29H23.83V125.68z M44.81,147.9c5.38,0,8.07-2.32,8.07-6.97c0-2.39-0.67-4.16-2-5.31\n\t\t\t c-1.33-1.15-3.36-1.73-6.07-1.73H34.59v14.01H44.81z\" />\n <path class=\"product-documents__svg-pdf\" d=\"M69.92,125.68h18.91c5.29,0,9.84,0.97,13.66,2.9c3.82,1.93,6.74,4.72,8.76,8.35\n\t\t\t c2.02,3.63,3.04,7.98,3.04,13.04c0,5.06-1,9.42-3,13.08c-2,3.66-4.91,6.45-8.73,8.38c-3.82,1.93-8.4,2.9-13.73,2.9H69.92V125.68z\n\t\t\t M88.07,165.63c10.35,0,15.52-5.22,15.52-15.66c0-10.4-5.17-15.59-15.52-15.59h-7.38v31.26H88.07z\" />\n <path class=\"product-documents__svg-pdf\"\n d=\"M122.57,125.68h32.84v8.49h-22.22v11.18h20.84v8.49h-20.84v20.49h-10.63V125.68z\" />\n </g>\n </svg>\n <span class=\"product-documents__info\">Technical Datasheet</span>\n </a>\n </div>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel current\">\n <h3 id=\"bioz\" class=\"sub-title1\">Product References</h3>\n <div class=\"ProductDescriptionContainer product-droppdown__section-description-specific\">\n <object id=\"wobj-3835-13-0042-q\" type=\"text/html\"\n data=\"https://www.bioz.com/v_widget_6_0/3835/13-0042/?ex=1\" style=\"width: 100%; height: 193px\"></object>\n <div id=\"bioz-w-pb-3835-13-0042-q-div\" style=\"width: 100%\">\n <a id=\"bioz-w-pb-3835-13-0042-q\" style=\"font-size: 12px; text-decoration: none; color: #4698cb\"\n href=\"https://www.bioz.com/\" target=\"_blank\"><img src=\"https://cdn.bioz.com/assets/favicon.png\" style=\"\n width: 11px;\n height: 11px;\n vertical-align: baseline;\n padding-bottom: 0px;\n margin-left: 0px;\n margin-bottom: 0px;\n float: none;\n \" />\n Powered by Bioz</a>\n <a style=\"\n font-size: 12px;\n text-decoration: none;\n float: right;\n color: transparent;\n \" href=\"https://www.bioz.com/result/13-0042/product/EpiCypher/?cn=14-1048\" target=\"_blank\">\n See more details on Bioz</a>\n </div>\n </div>\n </div>\n </div>\n </div>\n </div>\n\n <script>\n $(document).ready(function () {\n var widget_micro_obj = new v_widget_obj('s', [1]);\n widget_micro_obj.request_catalog_number_widget_data_internal(\n '13-0042',\n '13-0042'\n );\n });\n </script>\n\n <style>\n .form-field-title .required-text {\n display: none !important;\n }\n\n .form-label {\n padding-left: 2rem;\n }\n\n .form-label-text {\n margin: 0;\n margin-left: 0 !important;\n }\n\n /* //////////// */\n\n /* BIOZ */\n td table {\n margin: 0;\n padding: 6px !important;\n }\n\n span.bioz-w-parent-hover {\n font-size: 15px;\n }\n\n td .bioz-w-tooltipx {\n padding-top: 0 !important;\n }\n\n /* .bioz-w-parent-hover:hover {\n text-decoration: underline white 2px !important;\n } */\n\n /* /////////// */\n\n .form-field-title .required-text {\n display: none !important;\n }\n\n .form-label {\n padding-left: 2rem;\n }\n\n .form-label-text {\n margin: 0;\n margin-left: 0 !important;\n }\n\n .bioz-w-header td {\n padding: 0;\n }\n\n .epicypher-table table {\n margin-right: auto;\n margin-left: auto;\n width: 100%;\n border-top: 1px solid lightgray;\n }\n\n .epicypher-table th {\n color: #3b3a48;\n }\n\n .epicypher-table td {\n color: #3b3a48;\n font-size: 0.9rem;\n }\n\n .epicypher-table td a {\n text-decoration: underline;\n }\n\n .epicypher-table td,\n th {\n border: 0.5px solid lightgray;\n text-align: left;\n padding: 8px;\n /* white-space: nowrap; */\n }\n\n .epicypher-table tr:nth-child(even) {\n background-color: #f9f9f9;\n }\n\n .epicypher-table td:first-child {\n border-left: 1px solid lightgray;\n }\n\n /* Tablet */\n\n @media only screen and (max-width: 1024px) {\n\n /* Force table to not be like tables anymore */\n .epicypher-table th {\n display: none !important;\n }\n\n /* BIOZ */\n td.bioz-w-tooltipx::before {\n display: none;\n }\n\n /* Hide table headers (but not display: none;, for accessibility) */\n .epicypher-table thead tr {\n position: absolute;\n top: -9999px;\n left: -9999px;\n }\n\n .epicypher-table tr {\n border: 1px solid #ccc;\n }\n\n .epicypher-table td {\n /* Behave like a \"row\" */\n /* border: none; */\n border-bottom: 1px solid #eee;\n position: relative;\n padding-left: 30%;\n white-space: inherit;\n }\n\n .epicypher-table td:before {\n /* Now like a table header */\n position: absolute;\n /* Top/left values mimic padding */\n top: 6px;\n left: 6px;\n width: 45%;\n padding-right: 10px;\n white-space: nowrap;\n font-weight: 700;\n }\n\n /*\n Label the data\n */\n .epicypher-table td:nth-of-type(1):before {\n content: 'Item';\n }\n\n td:nth-of-type(2):before {\n content: 'Cat. No.';\n }\n\n /* BIOZ */\n }\n\n @media screen and (max-width: 767px) {\n .custom-title-description {\n font-size: 19px !important;\n }\n\n .section-title {\n font-size: 19px !important;\n padding-left: 1rem;\n }\n\n }\n </style>","tags":[],"warranty":"","price":{"without_tax":{"formatted":"$105.00","value":105,"currency":"USD"},"tax_label":"Sales Tax"},"detail_messages":"","availability":"","page_title":"CUTANA™ Rabbit IgG CUT&RUN Negative Control Antibody","cart_url":"https://www.epicypher.com/cart.php","max_purchase_quantity":0,"mpn":null,"upc":null,"options":[],"related_products":[{"id":694,"sku":null,"name":"CUTANA™ pAG-MNase for ChIC/CUT&RUN Workflows","url":"https://www.epicypher.com/products/epigenetics-reagents-and-assays/cutana-pag-mnase-for-chic-cut-and-run-workflows","availability":"","rating":null,"brand":{"name":null},"category":["Chromatin Mapping","Chromatin Mapping/CUTANA™ ChIC / CUT&RUN Assays"],"summary":"\n \n \n Type: Nuclease\n \n \n Mol Wgt: 43.7 kDa\n \n \n \n ","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/694/689/Screen_Shot_2020-02-12_at_11.01.55_AM__17144.1581530752.png?c=2","alt":"CUTANA™ pAG-MNase for ChIC/CUT&RUN Workflows"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/694/689/Screen_Shot_2020-02-12_at_11.01.55_AM__17144.1581530752.png?c=2","alt":"CUTANA™ pAG-MNase for ChIC/CUT&RUN Workflows"}],"date_added":"12th Aug 2019","pre_order":false,"show_cart_action":true,"has_options":true,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":[{"id":1174,"name":"Internal Comment","value":"Excess in bottom of Venom"},{"id":1175,"name":"Internal Comment","value":"Bulk in Psylocke"}],"num_reviews":null,"weight":{"formatted":"0.01 LBS","value":0.01},"demo":false,"price":{"without_tax":{"currency":"USD","formatted":"$335.00","value":335},"price_range":{"min":{"without_tax":{"currency":"USD","formatted":"$335.00","value":335},"tax_label":"Sales Tax"},"max":{"without_tax":{"currency":"USD","formatted":"$1,295.00","value":1295},"tax_label":"Sales Tax"}},"tax_label":"Sales Tax"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=694"},{"id":986,"sku":null,"name":"CUTANA™ CUT&Tag Kit","url":"https://www.epicypher.com/products/epigenetics-kits-and-reagents/cutana-cut-and-tag-kit","availability":"","rating":null,"brand":{"name":null},"category":["Antibodies/CUTANA™ CUT&Tag Antibodies","Chromatin Mapping/CUTANA™ CUT&Tag Assays"],"summary":"\n \n \n \n \n ","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/986/1213/CTK20__46720.1713641110.png?c=2","alt":"CUTANA™ CUT&Tag Kit"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/986/1213/CTK20__46720.1713641110.png?c=2","alt":"CUTANA™ CUT&Tag Kit"}],"date_added":"8th Mar 2023","pre_order":false,"show_cart_action":true,"has_options":true,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":[{"id":1219,"name":"Pack Size","value":"48 Reactions"}],"num_reviews":null,"weight":{"formatted":"0.01 LBS","value":0.01},"demo":false,"price":{"without_tax":{"currency":"USD","formatted":"$2,695.00","value":2695},"tax_label":"Sales Tax"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=986"},{"id":764,"sku":null,"name":"CUTANA™ pAG-Tn5 for CUT&Tag","url":"https://www.epicypher.com/products/epigenetics-kits-and-reagents/cutana-pag-tn5-for-cut-and-tag","availability":"","rating":null,"brand":{"name":null},"category":["Chromatin Mapping","Chromatin Mapping/CUTANA™ CUT&Tag Assays"],"summary":"\n \n \n Type: Transposase\n \n \n Mol Wgt: 191 kDa\n \n \n \n ","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/764/750/2020_CUTTag_icon_RGB_Andy_L__93238.1592491196.png?c=2","alt":"CUTANA™ pAG-Tn5 for CUT&Tag"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/764/750/2020_CUTTag_icon_RGB_Andy_L__93238.1592491196.png?c=2","alt":"CUTANA™ pAG-Tn5 for CUT&Tag"}],"date_added":"17th Jun 2020","pre_order":false,"show_cart_action":true,"has_options":true,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":[{"id":1005,"name":"Internal Comment","value":"extra boxes in bottom shelf of Venom"}],"num_reviews":null,"weight":{"formatted":"0.01 LBS","value":0.01},"demo":false,"price":{"without_tax":{"currency":"USD","formatted":"$795.00","value":795},"price_range":{"min":{"without_tax":{"currency":"USD","formatted":"$795.00","value":795},"tax_label":"Sales Tax"},"max":{"without_tax":{"currency":"USD","formatted":"$2,995.00","value":2995},"tax_label":"Sales Tax"}},"tax_label":"Sales Tax"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=764"}],"shipping_messages":[],"rating":0,"meta_keywords":"CUTANA™ Rabbit IgG CUT&RUN Negative Control Antibody, CUT&RUN, cleavage under targets and release using nuclease, cutandrun, cutana, chromatin immunocleavage, ChIC, 13-0042","show_quantity_input":1,"title":"CUTANA™ IgG Negative Control Antibody for CUT&RUN and CUT&Tag","gift_wrapping_available":false,"min_purchase_quantity":0,"customizations":[],"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/758/739/1579731877.1280.1280__61771.1592491196.png?c=2","alt":"CUTANA™ IgG Negative Control Antibody for CUT&RUN and CUT&Tag"}]} Pack Size: 100 µg
- Type: Polyclonal
- Host: Rabbit
- Applications: CUT&RUN, CUT&Tag
- Reactivity: Negative Control
- Format: Affinity Purified IgG
- Target Size: N/A
Description
Cleavage Under Targets & Release Using Nuclease (CUT&RUN) and Cleavage Under Targets and Tagmentation (CUT&Tag) are next generation assays for genomic mapping of chromatin features to replace Chromatin Immunoprecipitation (ChIP-seq) [Skene & Henikoff, 2017]. Unlike in ChIP-seq, where baseline signal is established by sequencing the fragmented input chromatin, reactions performed with this rabbit IgG antibody are used to determine background signal arising from non-specific MNase digestion and Tn5 tagmentation. Pair with the H3K4me3 positive control antibody (EpiCypher 13-0060) for a well-controlled experiment.
Validation Data - CUT&RUN
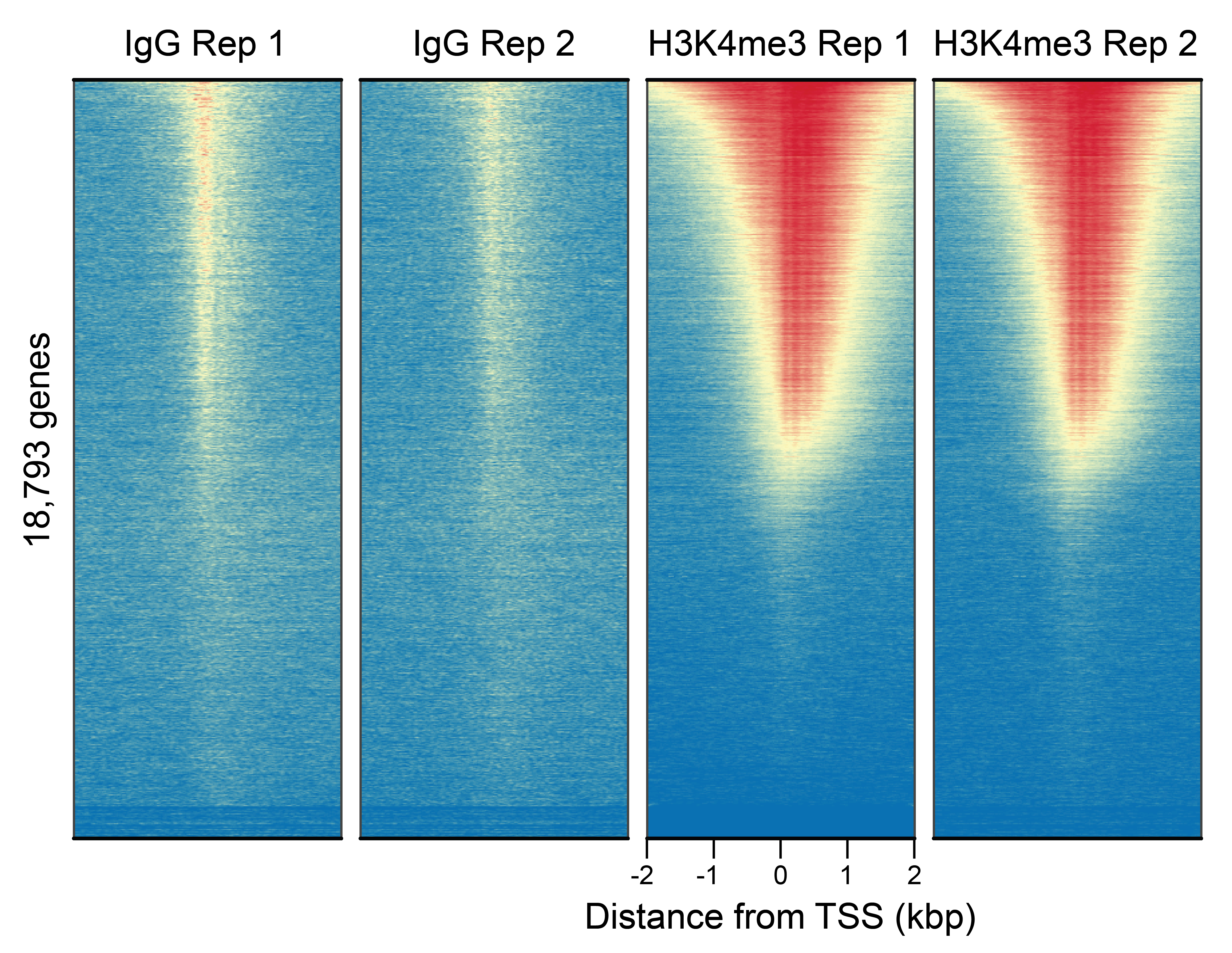
Figure 1: CUT&RUN genome wide
enrichment
CUT&RUN was performed as described in Figure 3. Heatmaps show H3K4me3 peaks
relative to IgG antibody in aligned rows
ranked by intensity (top to bottom) and colored such that red indicates high localized enrichment
and blue denotes
background signal. H3K4me3 antibody showed expected enrichment around the TSS. Despite some
observable background in the
IgG control, differential enrichment with the target antibody is as expected.
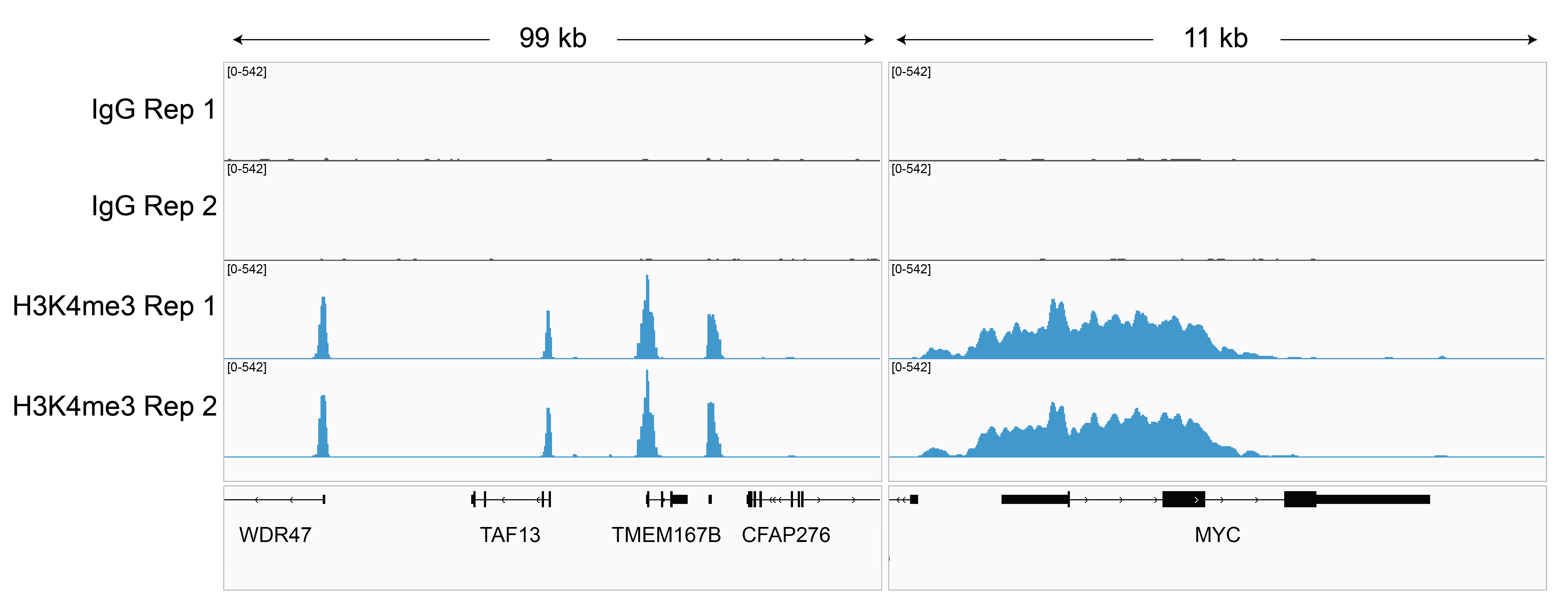
Figure 2: Rabbit IgG CUT&RUN tracks
CUT&RUN was performed as described in Figure 3. Gene browser shots were generated
using the Integrative Genomics Viewer (IGV, Broad Institute). Two gene loci show H3K4me3 peaks and
minimal background in the IgG track, as expected.
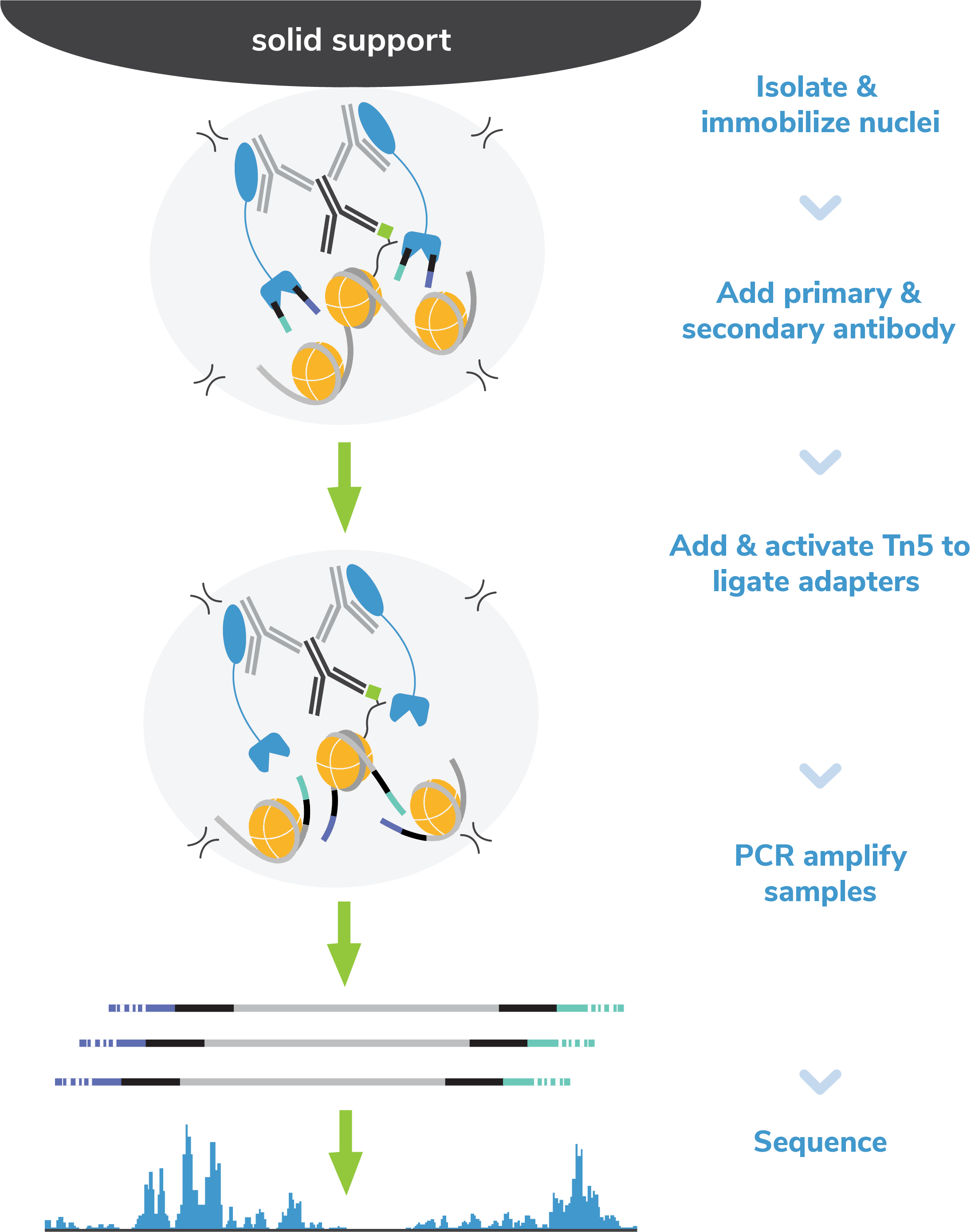
Figure 3: CUT&RUN methods
CUT&RUN was performed on 500k native K562 cells with 0.5 µg of either IgG or H3K4me3 (EpiCypher
13-0060) antibodies in duplicate using the CUTANA™ ChIC/CUT&RUN Kit v4 (EpiCypher 14-1048). Library
preparation was performed with 5 ng of DNA (or the total amount recovered if less than 5 ng) using
the CUTANA™ CUT&RUN Library Prep Kit (EpiCypher 14-1001/14-1002). Both kit protocols were adapted
for high throughput Tecan liquid handling. Sample sequencing depth was 7.8/6.8 million reads (IgG
Rep 1/Rep 2) and 12.6/10.9 million reads (H3K4me3 Rep 1/Rep 2). Data were aligned to the hg19 genome
using Bowtie2. Data were filtered to remove duplicates, multi-aligned reads, and ENCODE DAC
Exclusion List regions.
Validation Data - CUT&Tag
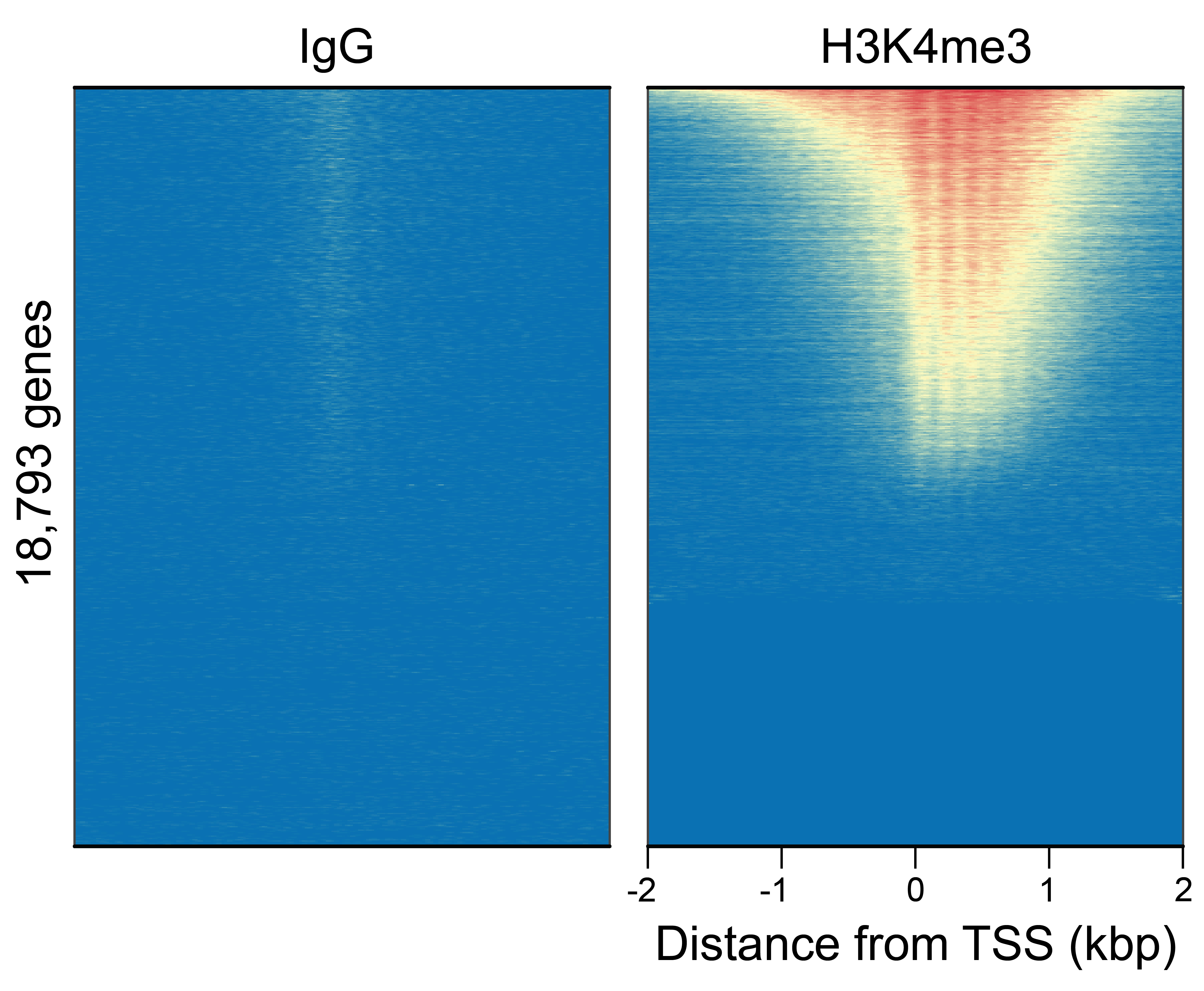
Figure 4: CUT&Tag genome wide
enrichment
CUT&Tag was performed as described in Figure 6. Heatmaps show H3K4me3 peaks
relative to IgG antibody in aligned rows ranked by intensity (top to bottom) and colored such that
red indicates high localized enrichment and blue denotes background signal. H3K4me3 antibody showed
expected enrichment around the TSS, while the IgG antibody displayed minimal background, as
expected.
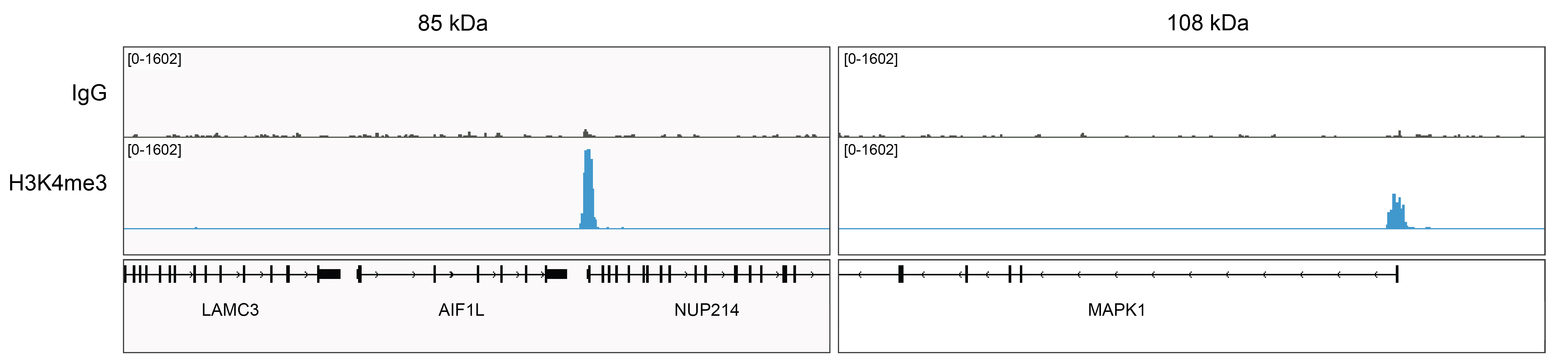
Figure 5: Rabbit IgG CUT&Tag tracks
CUT&Tag was performed as described in Figure 6. Gene browser shots were generated
using the Integrative Genomics Viewer (IGV, Broad Institute). Two gene loci show H3K4me3 peaks and
minimal background in the IgG track, as expected.
Figure 6: CUT&Tag methods
CUT&Tag was performed on 100k native K562 nuclei with 0.5 µg of either IgG or H3K4me3 (EpiCypher 13-0041) antibodies in singlicate using the CUTANA™ CUT&Tag Kit v1 (EpiCypher
14-1102/14-1103). Libraries were run on an Illumina NextSeq2000 with
paired-end sequencing (2x50bp). Sample sequencing depth was 0.9 million reads (IgG) and 2.9 million
reads (H3K4me3). Data were aligned to the hg19 genome using Bowtie2. Data were filtered to remove
duplicates, multi-aligned reads, and ENCODE DAC Exclusion List regions.