CUT&RUN and CUT&Tag Growth Outpacing ATAC-seq

Cleavage Under Targets and Release Using Nuclease (CUT&RUN) is an innovative chromatin mapping strategy, which leverages breakthroughs from the Chromatin ImmunoCleavage (ChIC) technique2 to selectively fragment antibody-bound chromatin loci for downstream sequencing. CUT&RUN was developed by Dr. Steven Henikoff’s laboratory at the Fred Hutchison Cancer Research Center, and was first published in 20173. The recently released sister technology, CUT&Tag (Cleavage Under Targets and Tagmentation), was also developed by Dr. Henikoff and collaborators at Fred Hutch4. Together, these two approaches are driving massive advancements in chromatin profiling, as they generate high quality data using a fraction of the cellular input, sequencing depth, and labor/time compared to standard chromatin immunoprecipitation (ChIP-seq) assays.
EpiCypher has spent the last two years perfecting these technologies for reliable and robust chromatin mapping experiments. These efforts have resulted in the launch of our CUTANA platform and release of the key enzymes needed for each assay, namely pAG-MNase for CUT&RUN and pAG-Tn5 for CUT&Tag. Furthermore, we offer a complete CUTANA™ CUT&RUN Kit, a collection of CUTANA CUT&RUN Compatible Antibodies, and user-friendly CUT&RUN / Tag protocols that can be applied to map diverse chromatin targets.
In developing the CUTANA product line, we have had time to consider what makes CUT&RUN / CUT&Tag special, and why the field is in need of new mapping tools. Here, we take a look at the rapid rise in publications using CUT&RUN compared to another recently developed (and now widely implemented) next-generation sequencing technology, ATAC-seq. We discuss the advantages of profiling unique histone PTMs / proteins using CUT&RUN, and how these data provide additional context to the study of chromatin-regulated gene expression.
CUT&RUN Adoption Outpaces ATAC-seq
ATAC-seq (Assay for Transposase Accessible Chromatin) was first published in 2013 as a novel and high-throughput method to map “open” (i.e. accessible) chromatin genome-wide5. Compared to its predecessors, such as MNase-seq and DNase-seq, ATAC-seq generates data with greater sensitivity while also providing a rapid protocol compatible with reduced cell inputs. As a result, it has been implemented in labs worldwide to study chromatin structure, and is routinely paired with RNA-seq for analysis of gene regulation processes. Indeed, ATAC-seq is widely regarded as a transformative technology that has made chromatin accessibility a keystone of many genomics / epigenomics studies.
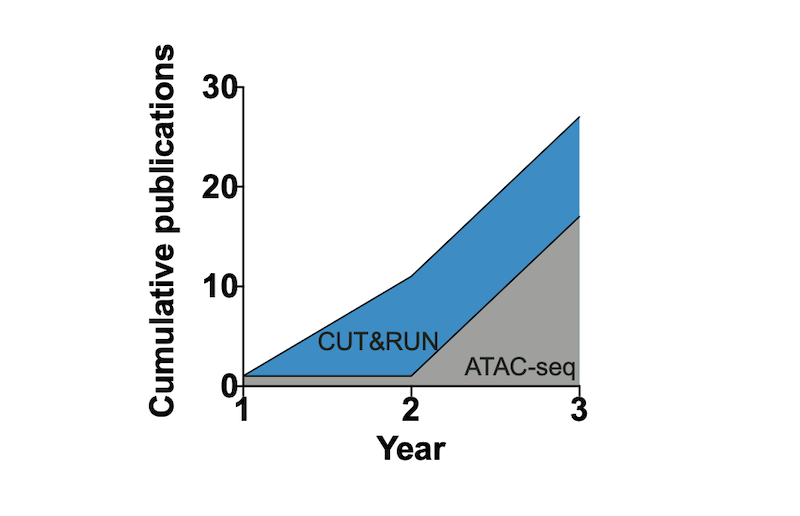
Figure 1: CUT&RUN adoption outpaces ATAC-seq. The graph shows the rapid publication rate in the first three years followed initial publication for CUT&RUN (2017) compared to ATAC-seq (2013). Publication numbers sourced from PubMed search results.
So how does the early adoption of CUT&RUN compare to ATAC-seq? The graph above illustrates the publication rate in the first three years following initial publication for CUT&RUN (20173) compared to ATAC-seq (20135). Shockingly, the application of CUT&RUN appears to be outpacing the now widely used ATAC-seq. Although it is difficult to make direct conclusions based on these data, it suggests that the chromatin field was eager for new and ultra-sensitive mapping techniques, and that many scientists are willing to try new strategies as part of their research projects.
Indeed, with many of the technical and cost limitations of ChIP-seq resolved by both CUT&RUN and CUT&Tag, we believe this trend will continue (and likely increase) for many years to come. For instance, CUT&RUN provides access to many chromatin-protein interactions inaccessible by conventional ChIP-seq analysis, particularly chromatin remodeling complexes, such as the SMARCA4 ATPase (Figure 2). In fact, EpiCypher has developed robust CUT&RUN protocols that can be used to profile a diverse array of chromatin features, including histone PTMs, transcription factors, chromatin remodelers, readers, and writer /eraser enzymes (Figure 2).
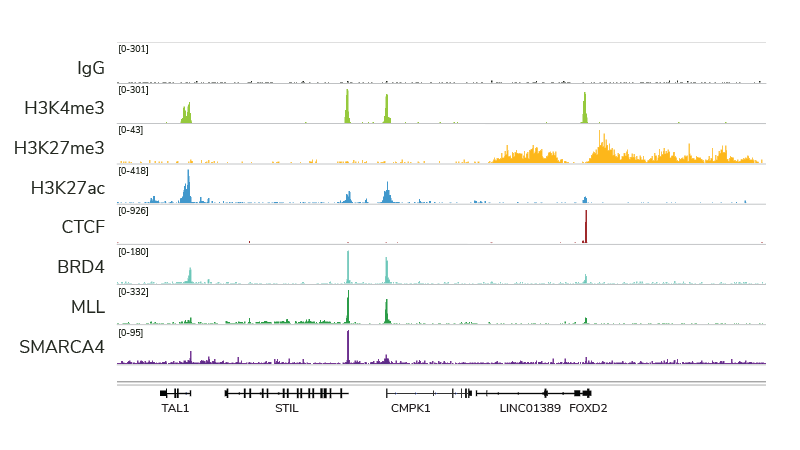
Figure 2: CUT&RUN profiling provides robust data for a variety of targets. The figure provides example data tracks, each generated using 500,000 K562 cells and 3-5 million paired end reads.
We’ve covered the advantages of CUT&RUN and CUT&Tag vs. ChIP-seq in a previous post, and have summarized these benefits below in Table 1. Aside from the increased target diversity, CUT&RUN / Tag provide massive improvements in signal-to-noise and background, while at the same time requiring drastically fewer cells and sequencing reads compared to ChIP-seq3,4,6-8. This means that more samples can be multiplexed for sequencing, allowing you to run more replicates / timepoints or simply save on costs; we estimate that total CUT&RUN assay costs (including library preparation and sequencing) are less than $100 / sample.
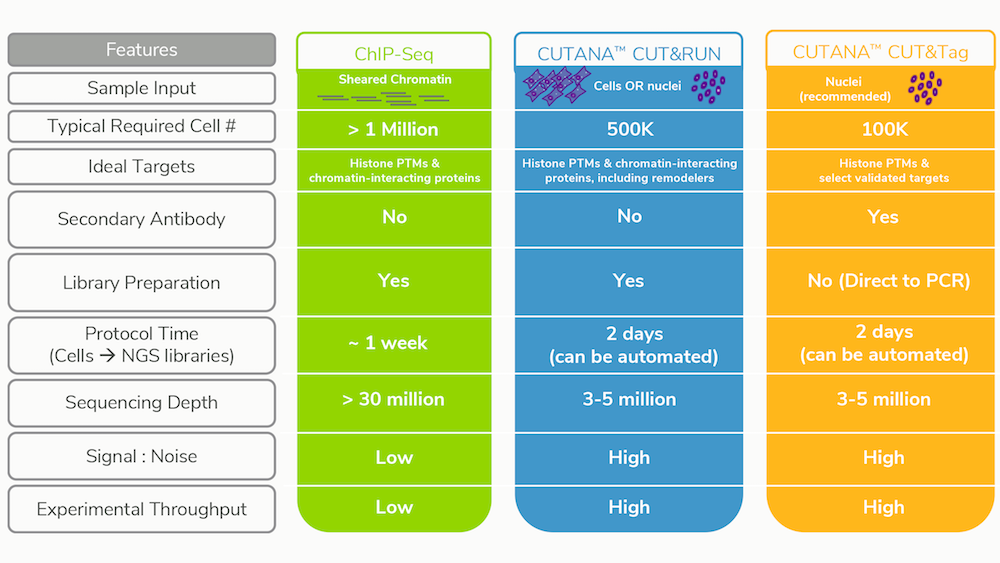
Table 1: Improvements and features of CUT&RUN and CUT&Tag vs. conventional
ChIP-seq.
*Cells are also compatible with CUT&Tag, however, tagmented
mitochondrial DNA will contribute to sequencing reads.
**Compatible with
benchtop sequencers for rapid results, thus democratizing genomics studies.
CUT&RUN / CUT&Tag Enable Comprehensive Gene Regulation Studies
As noted above, many scientists studying chromatin regulation of gene expression perform parallel RNA-seq and ATAC-seq experiments, thus providing a genome-wide view of transcription and chromatin accessibility, respectively. However, it is crucial to note that the terms “open” and “closed” chromatin are not binary. Indeed, within these two subsets there are complex chromatin signatures, each of which have distinct impacts on gene expression.
Figure 3 outlines this idea by comparing the accessible regions defined by ATAC-seq (top row) with the localization of unique chromatin features, such as histone PTMs, DNA methylation, and other chromatin associated proteins (ChAPs). Within the open chromatin regions identified in ATAC-seq, there are enhancers, H3K4me3-denoted “active” promoters, and promoters activated by other chromatin bound factors, such as transcription factors. The same is true for heterochromatin, as these areas can be described as more permanently repressed (i.e. “constitutive” heterochromatin, marked by H3K9me3) or as “facultative,” meaning that it has the ability to become accessible to transcriptional machinery (marked by H3K27me3).
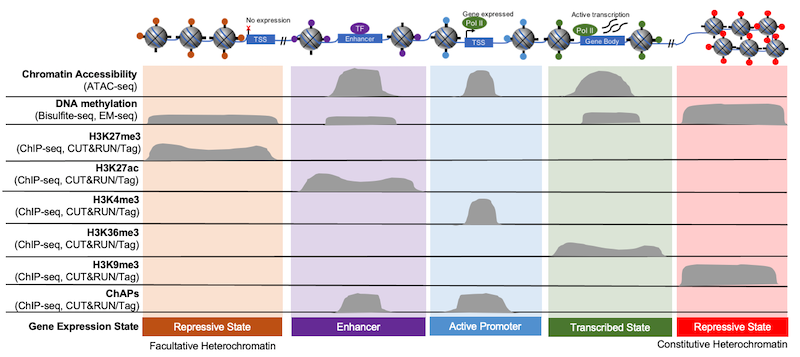
Figure 3: Gene expression is regulated by chromatin structure and accessibility. Histone PTMs provide useful markers of various chromatin elements, such as promoters, enhancers, and gene bodies. Adapted from 1.
Understandably, many researchers began using ATAC-seq as a universal chromatin accessibility assay, in lieu of having to perform ChIP-seq for multiple PTMs / proteins. This provided massive savings on time and sequencing costs and streamlined research projects. But by only using ATAC-seq, researchers miss the rich complexity of the epigenome and lack the context of chromatin features that are driving gene expression in their studies. As such, it is incredibly challenging to examine mechanisms using only ATAC-seq and RNA-seq. With CUT&RUN and CUT&Tag technology, scientists can now integrate comprehensive chromatin mapping strategies into their gene regulatory studies, without sacrificing the time and expenses associated with performing multiple ChIP-seq and ATAC-seq assays.
Nevertheless, it is often useful to generate chromatin accessibility maps, either as preliminary data or as part of a larger, systems-wide characterization of chromatin state. To this end, the Henikoff lab has published a recent preprint demonstrating the application of H3K4me2/3 CUT&Tag assays for chromatin accessibility9. Compared to standard ATAC-seq, the CUT&Tag ATAC method required fewer reads, had lower rates of mitochondrial DNA contamination, and displayed higher signal-to-noise (as determined by Fraction of Reads in Peaks, or FRiP score10). This strategy would make it possible to generate chromatin accessibility maps and histone PTM / factor profiles using the same experimental protocol, thus enabling streamlined sample comparisons.
EpiCypher has been at the forefront of optimizing CUT&RUN / Tag products and protocols, and we offer an array of products to help get you started. Each of these products is lot-tested in CUT&RUN and/or CUT&Tag experiments, ensuring high-quality performance.
|
CUTANA ChIC/CUT&RUN Products |
CUTANA ChIC/CUT&Tag Products |
|
CUTANA Accessory Reagents |
Protocols |
References
- Jiang S et al. Integrating ChIP-seq with other functional genomics data. Brief Funct Genomics 17, 104-15 (2018). PubMed PMID: 29579165.
- Schmid M et al. ChIC and ChEC; genomic mapping of chromatin proteins. Mol Cell 16, 147-57 (2004). PubMed PMID: 15469830.
- Skene PJ et al. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. Elife 6, (2017). PubMed PMID: 28079019.
- Kaya-Okur HS et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat Commun 10, 1930 (2019). PubMed PMID: 31036827.
- Buenrostro JD et al. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10, 1213-8 (2013). PubMed PMID: 24097267.
- Kaya-Okur HS et al. Efficient low-cost chromatin profiling with CUT&Tag. Nat Protoc 15, 3264-83 (2020). PubMed PMID: 32913232.
- Meers MP et al. Improved CUT&RUN chromatin profiling tools. Elife 8, (2019). PubMed PMID: 31232687.
- Skene PJ et al. Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat Protoc 13, 1006-19 (2018). PubMed PMID: 29651053.
- Henikoff S et al. Efficient transcription-coupled chromatin accessibility mapping in situ. bioRxiv (2020). Full preprint available here.
