CHD1 CUTANA™ CUT&RUN Antibody
In stock
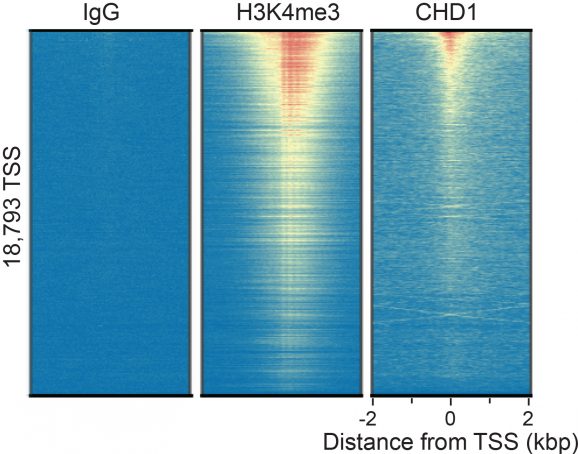
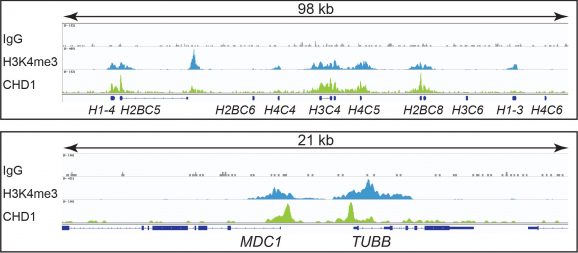
This antibody meets EpiCypher’s “CUTANA Compatible” criteria for performance in Cleavage Under Targets and Release Using Nuclease (CUT&RUN) and/or Cleavage Under Targets and Tagmentation (CUT&Tag) approaches to genomic mapping. Every lot of a CUTANA Compatible antibody is tested in the indicated approach using EpiCypher optimized protocols and determined to yield peaks that show a genomic distribution pattern consistent with reported function(s) of the target protein. CHD1 antibody produces CUT&RUN peaks above background (Figure 1) that overlap with H3K4me3 (Figures 1-2), consistent with CHD1 tandem chromodomains that mediate H3K4me3 binding and SWI/SNF complex nucleosome remodeling [1].
Trademarks and copyrights
Background References:
[1] Flanagan et al. Nature (2005). PMID: 16372014
CUT&RUN was performed on 500k K562 cells with either CHD1 (0.1 µg), H3K4me3 positive control (EpiCypher 13-0041; 0.5 µg), or IgG negative control (EpiCypher 13-0042; 0.5 µg) antibodies using the CUTANA™ ChIC/CUT&RUN Kit v2.0 (EpiCypher 14-1048). Library preparation was performed with 5 ng of DNA (or the total amount recovered if less than 5 ng) using the CUTANA™ CUT&RUN Library Prep Kit (EpiCypher 14-1001/14-1002). Both kit protocols were adapted for high throughput Tecan liquid handling. Libraries were run on an Illumina NextSeq2000 with paired-end sequencing (2×50 bp). Sample sequencing depth was 2.4 million reads (IgG), 4.1 million reads (H3K4me3), and 4.9 million reads (CHD1). Data were aligned to the hg19 genome using Bowtie2. Data were filtered to remove duplicates, multi-aligned reads, and ENCODE DAC Exclusion List regions.
Figure 1: CHD1 peaks in CUT&RUN
Figure 2: CHD1 CUT&RUN representative browser tracks
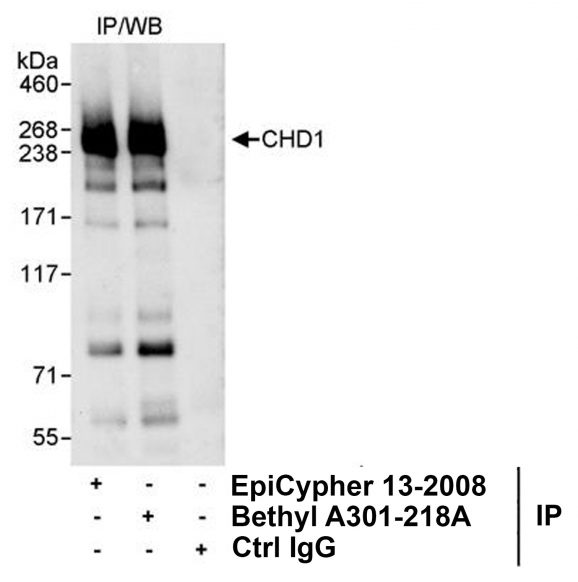
Figure 3: Immunoprecipitation data
- Type: Polyclonal
- Host: Rabbit
- Applications: CUT&RUN, IP
- Reactivity: Human
- Format: Antigen affinity-purified
- Target Size: 197 kDa
- CUT&RUN: 0.5 µg per reaction
- Immunoprecipitation (IP): 2 – 5 µg/mg lysate
- UniProt ID: O14646
- Gene Name: CHD1
- Protein Name: Chromodomain-helicase-DNA-binding protein 1
- Target Size: 197 kDa
- Alternate Names: ATP-dependent helicase CHD1, CHD-1