SP1 CUTANA™ CUT&RUN Antibody
{"url":"https://www.epicypher.com/products/antibodies/cutana-cut-run-antibodies/sp1-cutana-cut-run-antibody","add_this":[{"service":"facebook","annotation":""},{"service":"email","annotation":""},{"service":"print","annotation":""},{"service":"twitter","annotation":""},{"service":"linkedin","annotation":""}],"gtin":null,"id":908,"bulk_discount_rates":[],"can_purchase":true,"meta_description":"Rabbit polyclonal SP1 antibody rigorously tested for robust and reliable performance in CUT&RUN assays. Also validated for IHC, IP, & WB.","category":["Chromatin Mapping/CUTANA™ ChIC / CUT&RUN Assays","Antibodies/CUTANA™ CUT&RUN Antibodies","Antibodies/CUTANA™ CUT&RUN Antibodies/CUT&RUN Antibodies - Chromatin-Associated Proteins"],"AddThisServiceButtonMeta":"","main_image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/908/1008/chd3-cutana-cutandrun-antibody__66975.1645734525__91145.1648666822__51084.1648670485.jpg?c=2","alt":"SP1 CUTANA™ CUT&RUN Antibody"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=908","shipping":{"calculated":true},"num_reviews":0,"weight":"0.01 LBS","custom_fields":[{"id":"1031","name":"Pack Size","value":"100 µL"}],"sku":"13-2024","description":"<div class=\"product-general-info\">\n <ul class=\"product-general-info__list-left\">\n <li class=\"product-general-info__list-item\">\n <strong>Type: </strong>Polyclonal\n </li>\n <li class=\"product-general-info__list-item\">\n <strong>Host: </strong>Rabbit\n </li>\n <li class=\"product-general-info__list-item\">\n <strong>Applications: </strong>CUT&RUN, IHC, IP, WB\n </li>\n </ul>\n <ul class=\"product-general-info__list-right\">\n <li class=\"product-general-info__list-item\">\n <strong>Reactivity: </strong>Human, Mouse\n </li>\n <li class=\"product-general-info__list-item\">\n <strong>Format: </strong>Antigen affinity-purified\n </li>\n <li class=\"product-general-info__list-item\">\n <strong>Target Size: </strong>81 kDa\n </li>\n </ul>\n</div>\n\n<div class=\"service_accordion product-droppdown\">\n <div class=\"container\">\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel current\">\n <h3 class=\"sub-title1\">Description</h3>\n <div\n class=\"ProductDescriptionContainer product-droppdown__section-description-specific\">\n <p>\n This antibody meets EpiCypher’s \"CUTANA Compatible\" criteria for\n performance in Cleavage Under Targets and Release Using Nuclease\n (CUT&RUN) and/or Cleavage Under Targets and Tagmentation (CUT&Tag)\n approaches to genomic mapping. Every lot of a CUTANA Compatible\n antibody is tested in the indicated approach using EpiCypher\n optimized <a href=\"/resources/protocols/\">protocols</a> and\n determined to yield peaks that show a genomic distribution pattern\n consistent with reported function(s) of the target protein. SP1 is\n the original member of the Sp1-like family of zinc-finger\n transcription factors. It binds GC-rich motifs with high affinity\n [1] and is involved in expressing genes related to cell growth,\n cell-cycle progression, survival, and tumorigenesis [2]. SP1\n antibody produces CUT&RUN peaks above background primarily in\n intronic and promoter regions (<strong>Figure 1</strong>) that\n overlap with known SP1 DNA-binding motifs (<strong>Figure 2</strong\n >).\n </p>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel current\">\n <h3 class=\"sub-title1\">Validation Data</h3>\n <div\n class=\"ProductDescriptionContainer product-droppdown__section-description-specific\">\n <p>\n CUT&RUN was performed on 500k HeLa cells with 0.5 µg of either SP1\n or IgG negative control (EpiCypher\n <a\n href=\"/products/nucleosomes/snap-cutana-spike-in-controls/cutana-rabbit-igg-cut-run-negative-control-antibody\"\n >13-0042</a\n >) antibodies using the CUTANA™ ChIC/CUT&RUN Kit v2.0 (EpiCypher\n <a\n href=\"/products/epigenetics-reagents-and-assays/cutana-chic-cut-and-run-kit\"\n >14-1048</a\n >). Library preparation was performed with 5 ng of DNA (or the total\n amount recovered if less than 5 ng) using the CUTANA™ CUT&RUN\n Library Prep Kit (EpiCypher\n <a\n href=\"/products/epigenetics-reagents-and-assays/cutana-cut-and-run-library-prep-kit\"\n >14-1001/14-1002</a\n >). Both kit protocols were adapted for high throughput Tecan liquid\n handling. Libraries were run on an Illumina NextSeq2000 with\n paired-end sequencing (2x50 bp). Sample sequencing depth was 5.0\n million reads (IgG) and 5.3 million reads (SP1). Data were aligned\n to the hg19 genome using Bowtie2. Data were filtered to remove\n duplicates, multi-aligned reads, and ENCODE DAC Exclusion List regions.\n </p>\n\n <section class=\"image-picker\">\n <div class=\"image-picker__left\">\n <div\n class=\"image-picker__main-content_active image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg\n class=\"image-picker__svg-left\"\n width=\"24\"\n height=\"24\"\n viewBox=\"0 0 24 24\">\n <path\n d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a\n href=\"/content/images/products/antibodies/13-2024-sp1-peaks.jpeg\"\n target=\"_blank\"\n class=\"image-picker__main-image-link\"\n ><img loading=\"lazy\"\n alt=\"13-2024-sp1-peaks\"\n src=\"/content/images/products/antibodies/13-2024-sp1-peaks.jpeg\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\"\n >(Click to enlarge)</span\n ></a\n >\n <button class=\"image-picker__right-arrow\">\n <svg\n class=\"image-picker__svg-right\"\n width=\"24\"\n height=\"24\"\n viewBox=\"0 0 24 24\">\n <path\n d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\"\n ><strong>Figure 1: SP1 peaks in CUT&RUN </strong><br />\n CUT&RUN was performed as described above. Peaks were called\n with MACS2. Heatmaps show SP1 peaks relative to IgG negative\n control antibody in aligned rows ranked by intensity (top to\n bottom) and colored such that red indicates high localized\n enrichment and blue denotes background signal\n (<strong>left</strong>). The number of peaks that fall into\n distinct classes of functionally annotated genomic regions\n are shown (<strong>right</strong>).\n </span>\n </p>\n </div>\n <div class=\"image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg\n class=\"image-picker__svg-left\"\n width=\"24\"\n height=\"24\"\n viewBox=\"0 0 24 24\">\n <path\n d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a\n href=\"/content/images/products/antibodies/13-2024-binding-motif-analysis.jpeg\"\n target=\"_blank\"\n class=\"image-picker__main-image-link\"\n ><img loading=\"lazy\"\n alt=\"13-2024-binding-motif-analysis\"\n src=\"/content/images/products/antibodies/13-2024-binding-motif-analysis.jpeg\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\"\n >(Click to enlarge)</span\n ></a\n >\n <button class=\"image-picker__right-arrow\">\n <svg\n class=\"image-picker__svg-right\"\n width=\"24\"\n height=\"24\"\n viewBox=\"0 0 24 24\">\n <path\n d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\"\n ><strong\n >Figure 2: SP1 transcription factor binding motif analysis\n in CUT&RUN </strong\n ><br />\n Sp1(Zf)/Promoter/Homer consensus motif, represented as a\n sequence logo position weight matrix, was the top called\n motif significantly enriched under SP1 CUT&RUN peaks\n (<strong>left</strong>). The number of SP1 peaks containing\n the consensus motif from the left panel is represented by a\n Venn Diagram (<strong>right</strong>).\n </span>\n </p>\n </div>\n <div class=\"image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg\n class=\"image-picker__svg-left\"\n width=\"24\"\n height=\"24\"\n viewBox=\"0 0 24 24\">\n <path\n d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a\n href=\"/content/images/products/antibodies/13-2024-representative-browser-tracks.jpeg\"\n target=\"_blank\"\n class=\"image-picker__main-image-link\"\n ><img loading=\"lazy\"\n alt=\"13-2024-representative-browser-tracks\"\n src=\"/content/images/products/antibodies/13-2024-representative-browser-tracks.jpeg\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\"\n >(Click to enlarge)</span\n ></a\n >\n <button class=\"image-picker__right-arrow\">\n <svg\n class=\"image-picker__svg-right\"\n width=\"24\"\n height=\"24\"\n viewBox=\"0 0 24 24\">\n <path\n d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\"\n ><strong\n >Figure 3: SP1 CUT&RUN representative browser tracks </strong\n ><br />\n CUT&RUN was performed as described above. Gene browser shots\n were generated using the Integrative Genomics Viewer (IGV,\n Broad Institute). Three representative loci of the top\n called peaks are shown.\n </span>\n </p>\n </div>\n <div class=\"image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg\n class=\"image-picker__svg-left\"\n width=\"24\"\n height=\"24\"\n viewBox=\"0 0 24 24\">\n <path\n d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a\n href=\"/content/images/products/antibodies/13-2024-immunohistochemistry-data.jpeg\"\n target=\"_blank\"\n class=\"image-picker__main-image-link\"\n ><img loading=\"lazy\"\n alt=\"13-2024-immunohistochemistry-data\"\n src=\"/content/images/products/antibodies/13-2024-immunohistochemistry-data.jpeg\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\"\n >(Click to enlarge)</span\n ></a\n >\n <button class=\"image-picker__right-arrow\">\n <svg\n class=\"image-picker__svg-right\"\n width=\"24\"\n height=\"24\"\n viewBox=\"0 0 24 24\">\n <path\n d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\"\n ><strong>Figure 4: Immunohistochemistry data </strong><br />\n FFPE sections of human breast carcinoma\n (<strong>left</strong>) and mouse renal cell carcinoma\n (<strong>right</strong>) using SP1 antibody at a dilution of\n 1:5,000.\n </span>\n </p>\n </div>\n <div class=\"image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg\n class=\"image-picker__svg-left\"\n width=\"24\"\n height=\"24\"\n viewBox=\"0 0 24 24\">\n <path\n d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a\n href=\"/content/images/products/antibodies/13-2024-immunoprecipitation-data.jpeg\"\n target=\"_blank\"\n class=\"image-picker__main-image-link\"\n ><img loading=\"lazy\"\n alt=\"13-2024-immunoprecipitation-data\"\n src=\"/content/images/products/antibodies/13-2024-immunoprecipitation-data.jpeg\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\"\n >(Click to enlarge)</span\n ></a\n >\n <button class=\"image-picker__right-arrow\">\n <svg\n class=\"image-picker__svg-right\"\n width=\"24\"\n height=\"24\"\n viewBox=\"0 0 24 24\">\n <path\n d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\"\n ><strong>Figure 5: Immunoprecipitation data </strong><br />\n EpiCypher SP1 antibody (6 µg) was used to immunoprecipitate\n whole cell lysates (0.5 - 1 mg, 20% of IP loaded) isolated\n from HeLa cells using NETN lysis buffer. A negative control\n IgG antibody and positive control antibodies targeting SP1\n (Bethyl Laboratories) were also used to demonstrate\n specificity of the IP. EpiCypher 13-2024 and Bethyl\n A300-134A-5 target the same epitope, while Bethyl\n A300-944A-1 targets a different epitope (between amino acids\n 735 and 785). For blotting immunoprecipitates, EpiCypher SP1\n antibody was used at a 1:1,000 dilution.\n </span>\n </p>\n </div>\n <div class=\"image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg\n class=\"image-picker__svg-left\"\n width=\"24\"\n height=\"24\"\n viewBox=\"0 0 24 24\">\n <path\n d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\" />\n </svg>\n </button>\n <a\n href=\"/content/images/products/antibodies/13-2024-western-blot-data.jpeg\"\n target=\"_blank\"\n class=\"image-picker__main-image-link\"\n ><img loading=\"lazy\"\n alt=\"13-2024-western-blot-data\"\n src=\"/content/images/products/antibodies/13-2024-western-blot-data.jpeg\"\n class=\"image-picker__main-image\" />\n <span class=\"image-picker__main-image-caption\"\n >(Click to enlarge)</span\n ></a\n >\n <button class=\"image-picker__right-arrow\">\n <svg\n class=\"image-picker__svg-right\"\n width=\"24\"\n height=\"24\"\n viewBox=\"0 0 24 24\">\n <path\n d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\" />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\"\n ><strong>Figure 6: Western blot data </strong><br />\n Western analysis of SP1 in whole cell extracts from HeLa,\n HEK293T, and mouse Renca cells. Fifty micrograms of lysate\n was resolved via SDS-PAGE and detected with 1:10,000\n dilution of SP1 antibody.\n </span>\n </p>\n </div>\n </div>\n <aside class=\"image-picker__right\">\n <div class=\"image-picker__gallery\">\n <img loading=\"lazy\"\n alt=\"13-2024-sp1-peaks\"\n src=\"/content/images/products/antibodies/13-2024-sp1-peaks.jpeg\"\n width=\"200\"\n class=\"image-picker__side-image image-picker__side-image_active\"\n role=\"button\" />\n <img loading=\"lazy\"\n alt=\"13-2024-binding-motif-analysis\"\n src=\"/content/images/products/antibodies/13-2024-binding-motif-analysis.jpeg\"\n width=\"200\"\n class=\"image-picker__side-image image-picker__side-image_active\"\n role=\"button\" />\n <img loading=\"lazy\"\n alt=\"13-2024-representative-browser-tracks\"\n src=\"/content/images/products/antibodies/13-2024-representative-browser-tracks.jpeg\"\n width=\"200\"\n class=\"image-picker__side-image image-picker__side-image_active\"\n role=\"button\" />\n <img loading=\"lazy\"\n alt=\"13-2024-immunohistochemistry-data\"\n src=\"/content/images/products/antibodies/13-2024-immunohistochemistry-data.jpeg\"\n width=\"200\"\n class=\"image-picker__side-image image-picker__side-image_active\"\n role=\"button\" />\n <img loading=\"lazy\"\n alt=\"13-2024-immunoprecipitation-data\"\n src=\"/content/images/products/antibodies/13-2024-immunoprecipitation-data.jpeg\"\n width=\"200\"\n class=\"image-picker__side-image image-picker__side-image_active\"\n role=\"button\" />\n <img loading=\"lazy\"\n alt=\"13-2024-western-blot-data\"\n src=\"/content/images/products/antibodies/13-2024-western-blot-data.jpeg\"\n width=\"200\"\n class=\"image-picker__side-image image-picker__side-image_active\"\n role=\"button\" />\n </div>\n </aside>\n </section>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Technical Information</h3>\n <div\n class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <div class=\"product-tech-info\">\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Immunogen</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n Between amino acids 750 and 785\n </div>\n </div>\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Storage</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n Stable for 1 year at 4°C from date of receipt\n </div>\n </div>\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Formulation</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n Antigen affinity-purified antibody in Tris-citrate/phosphate\n buffer pH 7-8, 0.09% sodium azide\n </div>\n </div>\n </div>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Recommended Dilution</h3>\n <div\n class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <!-- <p><strong>CUT&RUN</strong> 0.5 µg per reaction</p>\n <p>\n <strong>Immunohistochemistry (IHC):</strong> 1:2,000 - 1:10,000.\n Epitope retrieval with citrate buffer pH 6.0 is recommended for FFPE\n tissue sections\n </p>\n <p><strong>Immunoprecipitation (IP):</strong> 2 - 10 µg/mg lysate</p>\n <p><strong>Western Blot (WB):</strong> 1:2,000 - 1:10,000</p> -->\n\n <div class=\"product-tech-info\">\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>CUT&RUN:</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n 0.5 µg per reaction\n </div>\n </div>\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Immunohistochemistry (IHC):</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n 1:2,000 - 1:10,000. Epitope retrieval with citrate buffer pH 6.0\n is recommended for FFPE tissue sections\n </div>\n </div>\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Immunoprecipitation (IP):</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n 2 - 10 µg/mg lysate\n </div>\n </div>\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Western Blot (WB):</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n 1:2,000 - 1:10,000\n </div>\n </div>\n </div>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Gene & Protein Information</h3>\n <div\n class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <div class=\"product-tech-info\">\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>UniProt ID</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">P08047</div>\n </div>\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Gene Name</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">SP1</div>\n </div>\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Protein Name</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n Transcription factor SP1\n </div>\n </div>\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Target Size</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">81 kDa</div>\n </div>\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Alternate Names</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n specificity protein 1, transcription factor SP1, TSFP1\n </div>\n </div>\n </div>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">References</h3>\n <div\n class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <strong>Background References:</strong>\n <br />\n [1] Raiber et al. <em>Nucleic Acids Res</em> (2012). PMID:\n <a\n href=\"https://pubmed.ncbi.nlm.nih.gov/22021377/\"\n title=\"A non-canonical DNA structure is a binding motif for the transcription factor SP1 in vitro\"\n target=\"new\">\n 22021377</a\n ><br />\n [2] Spengler et al. <em>Cell Cycle</em> (2008). PMID:\n <a\n href=\"https://pubmed.ncbi.nlm.nih.gov/18239466/\"\n title=\"Phosphorylation mediates Sp1 coupled activities of proteolytic processing, desumoylation and degradation\"\n target=\"new\">\n 18239466</a\n ><br />\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Documents & Resources</h3>\n <div\n class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <div class=\"product-documents\">\n <a\n href=\"/content/documents/tds/13-2024.pdf\"\n target=\"_blank\"\n class=\"product-documents__link\">\n <svg\n version=\"1.1\"\n id=\"Layer_1\"\n xmlns=\"http://www.w3.org/2000/svg\"\n xmlns:xlink=\"http://www.w3.org/1999/xlink\"\n x=\"0px\"\n y=\"0px\"\n viewBox=\"0 0 228 240\"\n style=\"enable-background: new 0 0 228 240\"\n xml:space=\"preserve\"\n class=\"product-documents__icon\"\n alt=\"16-0030 Datasheet\">\n <g>\n <path\n class=\"product-documents__svg-pdf\"\n d=\"M191.92,68.77l-47.69-47.69c-1.33-1.33-3.12-2.08-5.01-2.08H45.09C41.17,19,38,22.17,38,26.09v184.36\n c0,3.92,3.17,7.09,7.09,7.09h141.82c3.92,0,7.09-3.17,7.09-7.09V73.8C194,71.92,193.25,70.1,191.92,68.77z M177.65,77.06h-41.7\n v-41.7L177.65,77.06z M178.05,201.59H53.95V34.95h66.92v47.86c0,5.14,4.17,9.31,9.31,9.31h47.86V201.59z\" />\n </g>\n <rect\n x=\"20\"\n y=\"112\"\n class=\"product-documents__svg-background\"\n width=\"146\"\n height=\"76\" />\n <g>\n <path\n class=\"product-documents__svg-pdf\"\n d=\"M23.83,125.68h22.36c5.29,0,9.41,1.33,12.35,4c2.94,2.67,4.42,6.39,4.42,11.18c0,4.78-1.47,8.51-4.42,11.18\n c-2.94,2.67-7.06,4-12.35,4H34.59v18.29H23.83V125.68z M44.81,147.9c5.38,0,8.07-2.32,8.07-6.97c0-2.39-0.67-4.16-2-5.31\n c-1.33-1.15-3.36-1.73-6.07-1.73H34.59v14.01H44.81z\" />\n <path\n class=\"product-documents__svg-pdf\"\n d=\"M69.92,125.68h18.91c5.29,0,9.84,0.97,13.66,2.9c3.82,1.93,6.74,4.72,8.76,8.35\n c2.02,3.63,3.04,7.98,3.04,13.04c0,5.06-1,9.42-3,13.08c-2,3.66-4.91,6.45-8.73,8.38c-3.82,1.93-8.4,2.9-13.73,2.9H69.92V125.68z\n M88.07,165.63c10.35,0,15.52-5.22,15.52-15.66c0-10.4-5.17-15.59-15.52-15.59h-7.38v31.26H88.07z\" />\n <path\n class=\"product-documents__svg-pdf\"\n d=\"M122.57,125.68h32.84v8.49h-22.22v11.18h20.84v8.49h-20.84v20.49h-10.63V125.68z\" />\n </g>\n </svg>\n <span class=\"product-documents__info\">Technical Datasheet</span>\n </a>\n </div>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Additional Info</h3>\n <div\n class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <p>\n This antibody is provided for commercial sale under license from\n Bethyl Laboratories, Inc.\n </p>\n </div>\n </div>\n </div>\n </div>\n</div>\n","tags":[],"warranty":"","price":{"without_tax":{"formatted":"$525.00","value":525,"currency":"USD"},"tax_label":"Sales Tax"},"detail_messages":"","availability":"","page_title":"SP1 CUTANA™ CUT&RUN Antibody","cart_url":"https://www.epicypher.com/cart.php","max_purchase_quantity":0,"mpn":null,"upc":null,"options":[],"related_products":[{"id":889,"sku":null,"name":"CUTANA™ CUT&RUN Library Prep Kit","url":"https://www.epicypher.com/products/epigenetics-kits-and-reagents/cutana-cut-run-library-prep-kit","availability":"","rating":null,"brand":{"name":null},"category":["Chromatin Mapping","Chromatin Mapping/CUTANA™ ChIC / CUT&RUN Assays"],"summary":"\n \n \n \n \n \n \n \n ","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/889/997/Epicypher_CUTANA_CUTRUN_LibraryPrep_Box__64431.1646406521.png?c=2","alt":"CUTANA™ CUT&RUN Library Prep Kit"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/889/997/Epicypher_CUTANA_CUTRUN_LibraryPrep_Box__64431.1646406521.png?c=2","alt":"CUTANA™ CUT&RUN Library Prep Kit"}],"date_added":"31st Jan 2022","pre_order":false,"show_cart_action":true,"has_options":true,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":[{"id":983,"name":"Pack Size","value":"48 Reactions"}],"num_reviews":null,"weight":{"formatted":"0.01 LBS","value":0.01},"demo":false,"price":{"without_tax":{"currency":"USD","formatted":"$1,525.00","value":1525},"tax_label":"Sales Tax"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=889"},{"id":694,"sku":null,"name":"CUTANA™ pAG-MNase for ChIC/CUT&RUN Workflows","url":"https://www.epicypher.com/products/epigenetics-reagents-and-assays/cutana-pag-mnase-for-chic-cut-and-run-workflows","availability":"","rating":null,"brand":{"name":null},"category":["Chromatin Mapping","Chromatin Mapping/CUTANA™ ChIC / CUT&RUN Assays"],"summary":"\n \n \n Type: Nuclease\n \n \n Mol Wgt: 43.7 kDa\n \n \n \n ","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/694/689/Screen_Shot_2020-02-12_at_11.01.55_AM__17144.1581530752.png?c=2","alt":"CUTANA™ pAG-MNase for ChIC/CUT&RUN Workflows"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/694/689/Screen_Shot_2020-02-12_at_11.01.55_AM__17144.1581530752.png?c=2","alt":"CUTANA™ pAG-MNase for ChIC/CUT&RUN Workflows"}],"date_added":"12th Aug 2019","pre_order":false,"show_cart_action":true,"has_options":true,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":[{"id":1174,"name":"Internal Comment","value":"Excess in bottom of Venom"},{"id":1175,"name":"Internal Comment","value":"Bulk in Psylocke"}],"num_reviews":null,"weight":{"formatted":"0.01 LBS","value":0.01},"demo":false,"price":{"without_tax":{"currency":"USD","formatted":"$335.00","value":335},"price_range":{"min":{"without_tax":{"currency":"USD","formatted":"$335.00","value":335},"tax_label":"Sales Tax"},"max":{"without_tax":{"currency":"USD","formatted":"$1,295.00","value":1295},"tax_label":"Sales Tax"}},"tax_label":"Sales Tax"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=694"},{"id":758,"sku":"13-0042","name":"CUTANA™ IgG Negative Control Antibody for CUT&RUN and CUT&Tag","url":"https://www.epicypher.com/products/nucleosomes/snap-cutana-spike-in-controls/cutana-igg-negative-control-antibody-for-cut-run-and-cut-tag","availability":"","rating":null,"brand":{"name":null},"category":["Nucleosomes/SNAP-CUTANA™ Spike-in Controls","Antibodies/CUTANA™ CUT&RUN Antibodies","Chromatin Mapping","Chromatin Mapping/CUTANA™ ChIC / CUT&RUN Assays","Chromatin Mapping/CUTANA™ CUT&Tag Assays"],"summary":"\n \n \n Type: Polyclonal\n \n \n Host: Rabbit\n \n \n Applications: CUT&R","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/758/739/1579731877.1280.1280__61771.1592491196.png?c=2","alt":"CUTANA™ IgG Negative Control Antibody for CUT&RUN and CUT&Tag"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/758/739/1579731877.1280.1280__61771.1592491196.png?c=2","alt":"CUTANA™ IgG Negative Control Antibody for CUT&RUN and CUT&Tag"}],"date_added":"17th Jun 2020","pre_order":false,"show_cart_action":true,"has_options":false,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":[{"id":1148,"name":"Pack Size","value":"100 µg"},{"id":1149,"name":"Internal Comment","value":"bulk stocks from Thermo in cold room"}],"num_reviews":null,"weight":{"formatted":"0.01 LBS","value":0.01},"demo":false,"add_to_cart_url":"https://www.epicypher.com/cart.php?action=add&product_id=758","price":{"without_tax":{"currency":"USD","formatted":"$105.00","value":105},"tax_label":"Sales Tax"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=758"}],"shipping_messages":[],"rating":0,"meta_keywords":"SP1 antibody, CUT&RUN antibody, transcription factor antibody, Sp1 transcription factor, Transcription factor Sp1, specificity protein 1, transcription factor Sp1, TSFP1","show_quantity_input":1,"title":"SP1 CUTANA™ CUT&RUN Antibody","gift_wrapping_available":false,"min_purchase_quantity":0,"customizations":[],"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/908/1008/chd3-cutana-cutandrun-antibody__66975.1645734525__91145.1648666822__51084.1648670485.jpg?c=2","alt":"SP1 CUTANA™ CUT&RUN Antibody"}]} Pack Size: 100 µL
- Type: Polyclonal
- Host: Rabbit
- Applications: CUT&RUN, IHC, IP, WB
- Reactivity: Human, Mouse
- Format: Antigen affinity-purified
- Target Size: 81 kDa
Description
This antibody meets EpiCypher’s "CUTANA Compatible" criteria for performance in Cleavage Under Targets and Release Using Nuclease (CUT&RUN) and/or Cleavage Under Targets and Tagmentation (CUT&Tag) approaches to genomic mapping. Every lot of a CUTANA Compatible antibody is tested in the indicated approach using EpiCypher optimized protocols and determined to yield peaks that show a genomic distribution pattern consistent with reported function(s) of the target protein. SP1 is the original member of the Sp1-like family of zinc-finger transcription factors. It binds GC-rich motifs with high affinity [1] and is involved in expressing genes related to cell growth, cell-cycle progression, survival, and tumorigenesis [2]. SP1 antibody produces CUT&RUN peaks above background primarily in intronic and promoter regions (Figure 1) that overlap with known SP1 DNA-binding motifs (Figure 2).
Validation Data
CUT&RUN was performed on 500k HeLa cells with 0.5 µg of either SP1 or IgG negative control (EpiCypher 13-0042) antibodies using the CUTANA™ ChIC/CUT&RUN Kit v2.0 (EpiCypher 14-1048). Library preparation was performed with 5 ng of DNA (or the total amount recovered if less than 5 ng) using the CUTANA™ CUT&RUN Library Prep Kit (EpiCypher 14-1001/14-1002). Both kit protocols were adapted for high throughput Tecan liquid handling. Libraries were run on an Illumina NextSeq2000 with paired-end sequencing (2x50 bp). Sample sequencing depth was 5.0 million reads (IgG) and 5.3 million reads (SP1). Data were aligned to the hg19 genome using Bowtie2. Data were filtered to remove duplicates, multi-aligned reads, and ENCODE DAC Exclusion List regions.
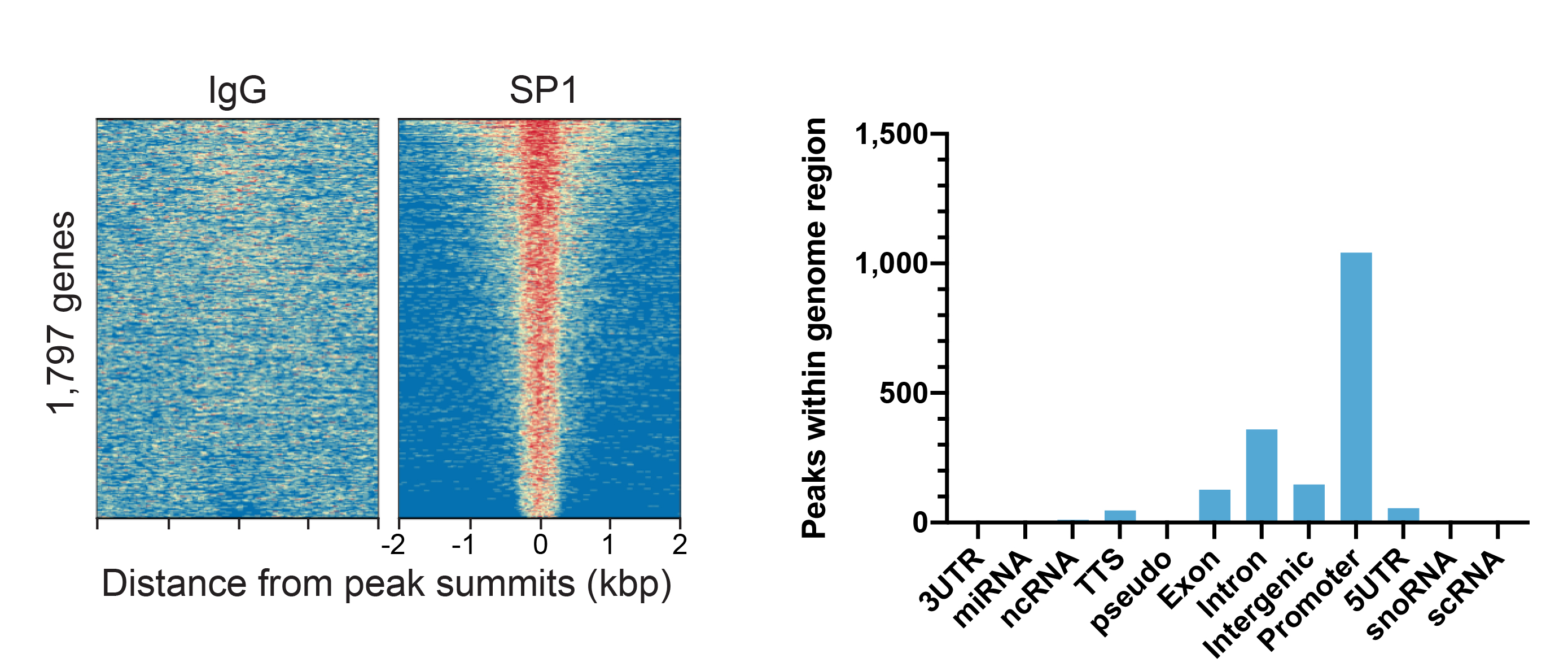
Figure 1: SP1 peaks in CUT&RUN
CUT&RUN was performed as described above. Peaks were called
with MACS2. Heatmaps show SP1 peaks relative to IgG negative
control antibody in aligned rows ranked by intensity (top to
bottom) and colored such that red indicates high localized
enrichment and blue denotes background signal
(left). The number of peaks that fall into
distinct classes of functionally annotated genomic regions
are shown (right).
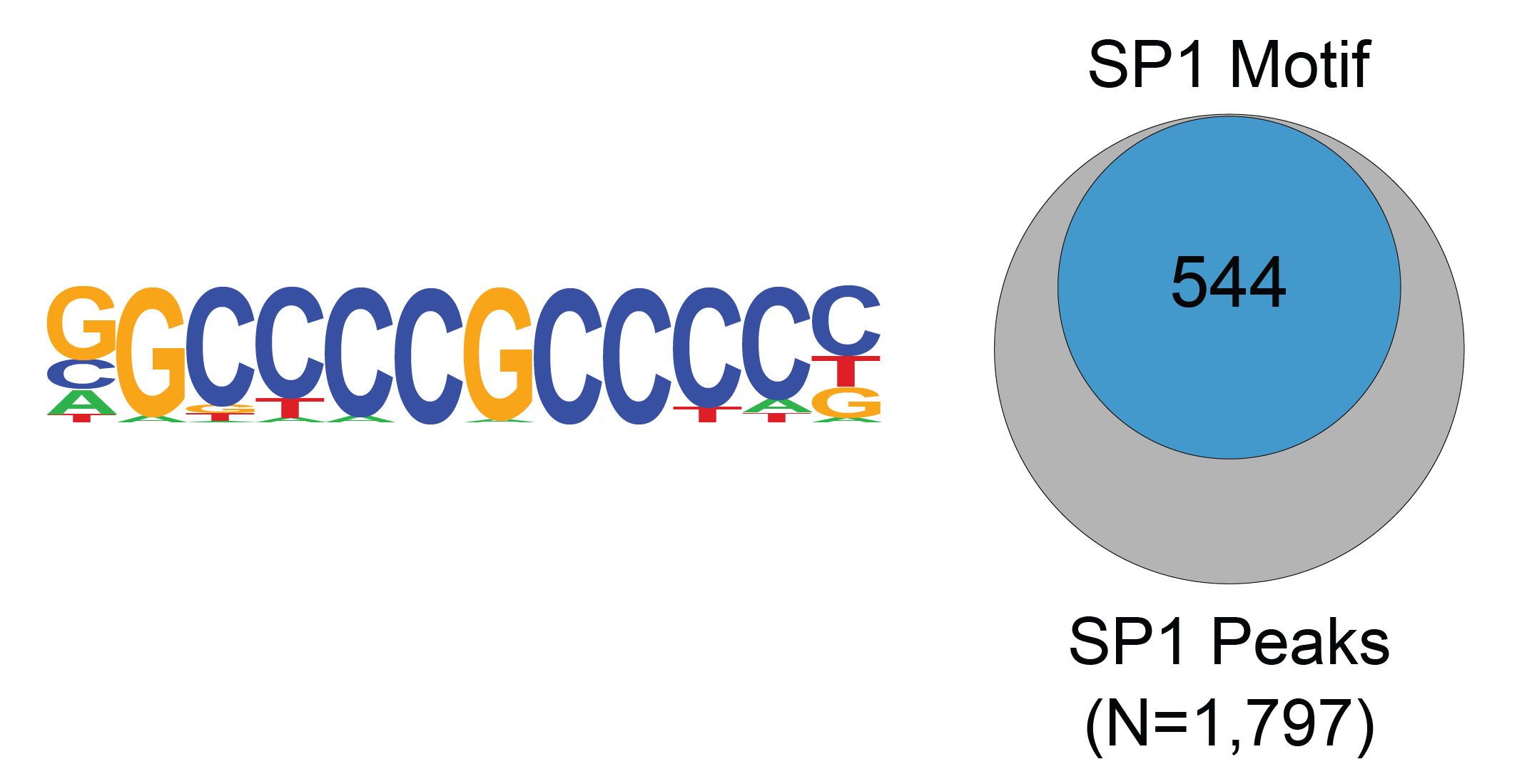
Figure 2: SP1 transcription factor binding motif analysis
in CUT&RUN
Sp1(Zf)/Promoter/Homer consensus motif, represented as a
sequence logo position weight matrix, was the top called
motif significantly enriched under SP1 CUT&RUN peaks
(left). The number of SP1 peaks containing
the consensus motif from the left panel is represented by a
Venn Diagram (right).
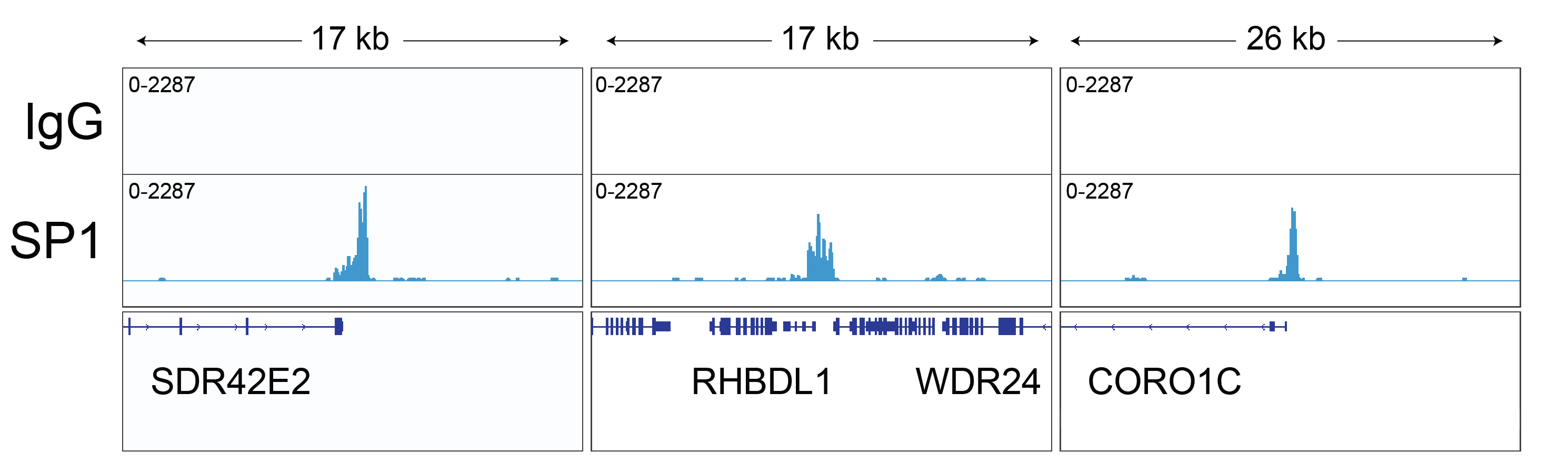
Figure 3: SP1 CUT&RUN representative browser tracks
CUT&RUN was performed as described above. Gene browser shots
were generated using the Integrative Genomics Viewer (IGV,
Broad Institute). Three representative loci of the top
called peaks are shown.
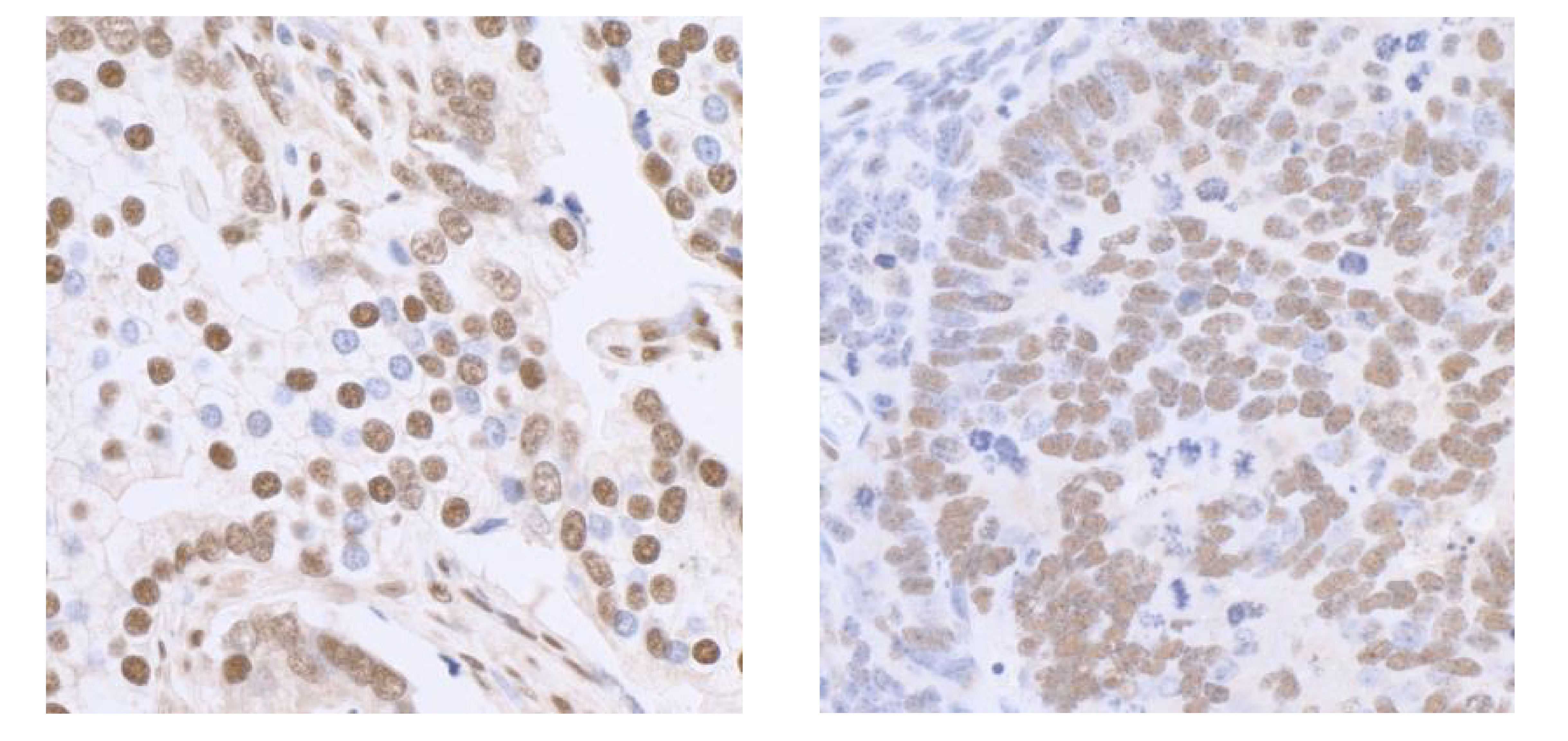
Figure 4: Immunohistochemistry data
FFPE sections of human breast carcinoma
(left) and mouse renal cell carcinoma
(right) using SP1 antibody at a dilution of
1:5,000.
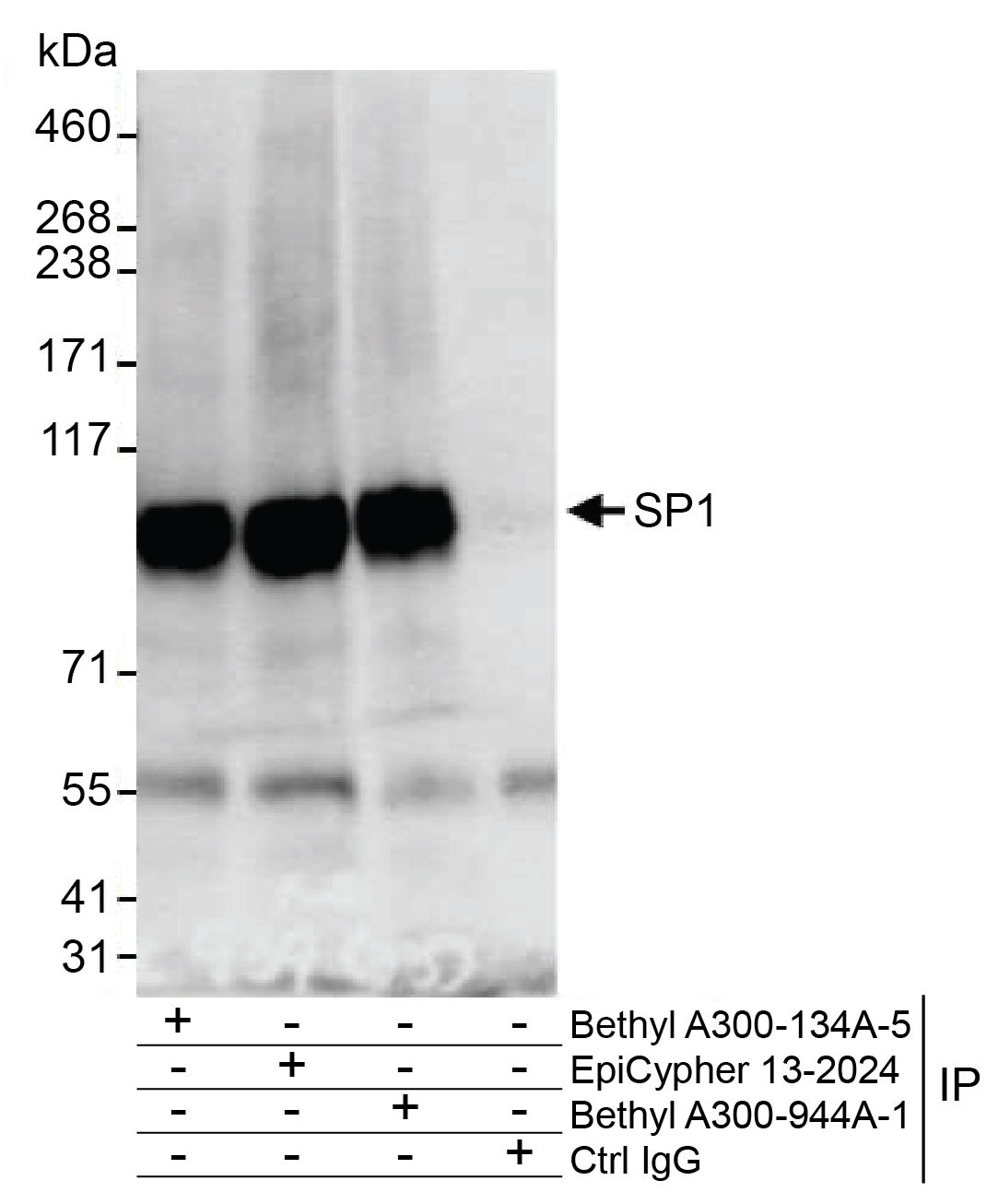
Figure 5: Immunoprecipitation data
EpiCypher SP1 antibody (6 µg) was used to immunoprecipitate
whole cell lysates (0.5 - 1 mg, 20% of IP loaded) isolated
from HeLa cells using NETN lysis buffer. A negative control
IgG antibody and positive control antibodies targeting SP1
(Bethyl Laboratories) were also used to demonstrate
specificity of the IP. EpiCypher 13-2024 and Bethyl
A300-134A-5 target the same epitope, while Bethyl
A300-944A-1 targets a different epitope (between amino acids
735 and 785). For blotting immunoprecipitates, EpiCypher SP1
antibody was used at a 1:1,000 dilution.
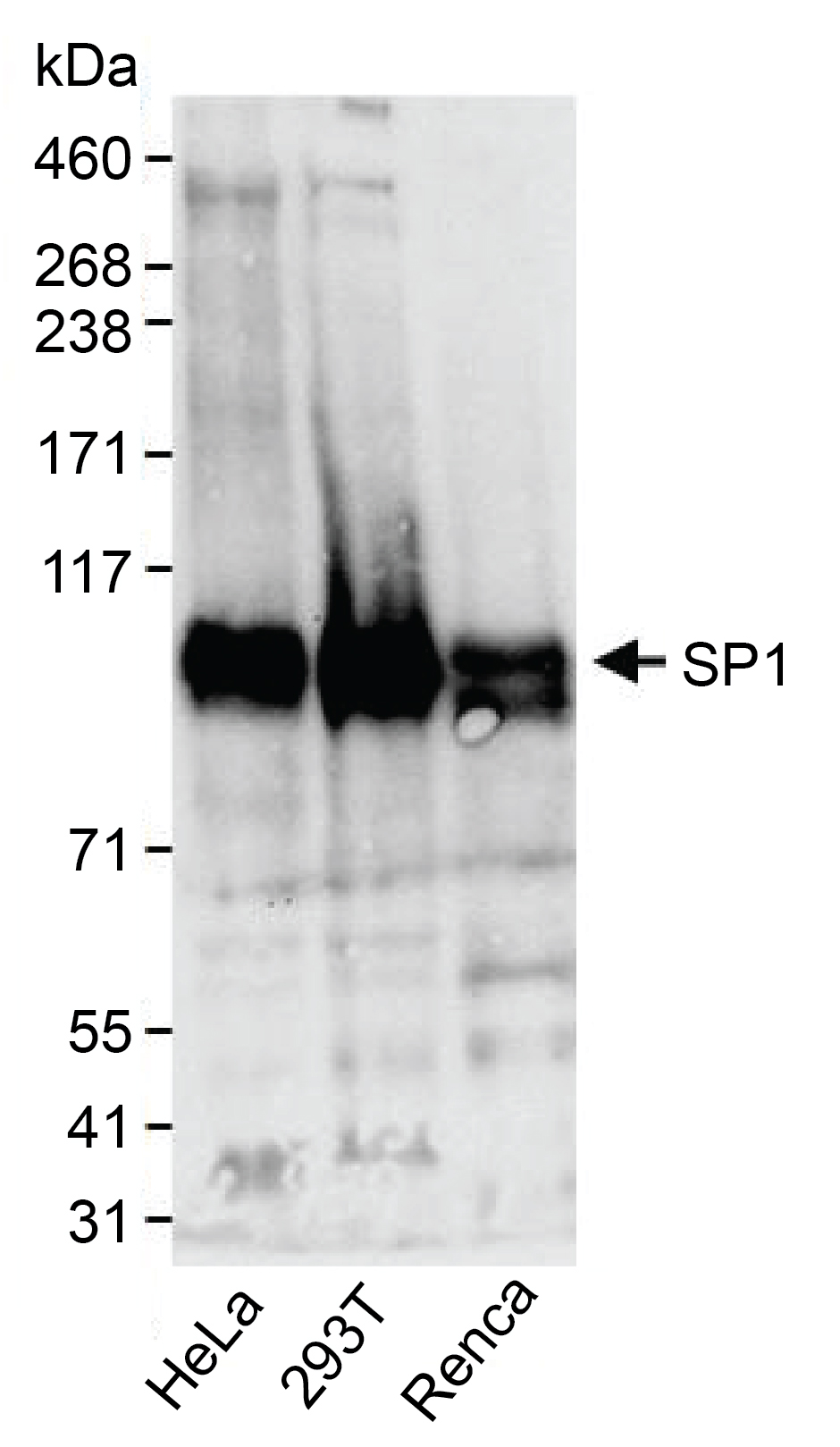
Figure 6: Western blot data
Western analysis of SP1 in whole cell extracts from HeLa,
HEK293T, and mouse Renca cells. Fifty micrograms of lysate
was resolved via SDS-PAGE and detected with 1:10,000
dilution of SP1 antibody.
Technical Information
Recommended Dilution
Gene & Protein Information
Documents & Resources
Additional Info
This antibody is provided for commercial sale under license from Bethyl Laboratories, Inc.