EpiDyne® Remodeling Assay Substrate DNA ST601-GATC0, Biotinylated
SKU: 18-4110
Description
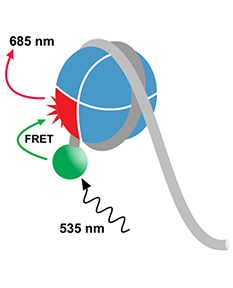
EpiDyne® Remodeling Assay Substrate DNA ST601-GATC0 is a
217 base-pair double-stranded DNA fragment containing the 601
nucleosome positioning sequence [1], which...
$115.00