EpiDyne® Remodeling Assay Substrate DNA ST601-GATC1
{"url":"https://www.epicypher.com/products/nucleosomes/epidyne-chromatin-remodeling-substrates/epidyne-remodeling-assay-substrate-dna-st601-gatc1","add_this":[{"service":"facebook","annotation":""},{"service":"email","annotation":""},{"service":"print","annotation":""},{"service":"twitter","annotation":""},{"service":"linkedin","annotation":""}],"gtin":null,"options":[],"id":374,"bulk_discount_rates":[],"can_purchase":true,"meta_description":"EpiDyne Remodeling Assay Substrate DNA (Positive Control)","category":["Nucleosomes/Nucleosome Components and Subunits","Nucleosomes/EpiDyne® Chromatin Remodeling Substrates"],"AddThisServiceButtonMeta":"","main_image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/374/292/DNA__93069.1614637188.gif?c=2","alt":"EpiDyne® Remodeling Assay Substrate DNA ST601-GATC1"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=374","shipping":{"calculated":true},"num_reviews":0,"weight":"0.01 LBS","custom_fields":[{"id":"637","name":"Pack Size","value":"50 µg"}],"sku":"18-4101","description":"<div class=\"service_accordion product-droppdown\">\n <div class=\"container\">\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel current\">\n <h3 class=\"sub-title1\">Description</h3>\n <div\n class=\"ProductDescriptionContainer product-droppdown__section-description-specific\"\n >\n <p>\n EpiDyne<sup>®</sup> Remodeling Assay Substrate DNA ST601-GATC1 is a\n 217 base-pair double-stranded DNA fragment containing the 601\n nucleosome positioning sequence [1], which has high affinity for\n histone octamers and is useful for nucleosome assembly. The DNA also\n includes a 3’ acceptor sequence to accommodate the histone octamer\n subsequent to remodeling. This positive control has a DpnII\n restriction site within the 601 sequence. When paired with the\n negative control DNA (EpiCypher\n <a\n href=\"/products/nucleosomes/epidyne-chromatin-remodeling-substrates/epidyne-remodeling-assay-substrate-dna-st601-gatc0\"\n >18-4100</a\n >) these controls illustrate the migration range for the Restriction\n Enzyme Assay. See the\n <a\n href=\"https://www.epicypher.com/content/documents/technotes/EpiDyne_REA.pdf\"\n >EpiDyne Nucleosome Remodeling Assay Tech Note</a\n >\n (Restriction Enzyme Assay) for more information.\n </p>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel current\">\n <h3 class=\"sub-title1\">Validation Data</h3>\n <div\n class=\"ProductDescriptionContainer product-droppdown__section-description-specific\"\n >\n <section class=\"image-picker\">\n <div class=\"image-picker__left\">\n <div\n class=\"image-picker__main-content_active image-picker__main-content\"\n >\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg\n class=\"image-picker__svg-left\"\n width=\"24\"\n height=\"24\"\n viewBox=\"0 0 24 24\"\n >\n <path\n d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\"\n />\n </svg>\n </button>\n <a\n href=\"/content/images/products/nucleosomes/18-4101-dna-gel-data.png\"\n target=\"_blank\"\n class=\"image-picker__main-image-link\"\n ><img\n alt=\"18-4101-dna-gel-data\"\n src=\"/content/images/products/nucleosomes/18-4101-dna-gel-data.png\"\n class=\"image-picker__main-image\"\n />\n <span class=\"image-picker__main-image-caption\"\n >(Click to enlarge)</span\n ></a\n >\n <button class=\"image-picker__right-arrow\">\n <svg\n class=\"image-picker__svg-right\"\n width=\"24\"\n height=\"24\"\n viewBox=\"0 0 24 24\"\n >\n <path\n d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\"\n />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\"\n ><strong>Figure 1: DNA gel data </strong><br />\n ST601-GATC1 DNA resolved via native PAGE gel and stained\n with ethidium bromide to visualize DNA.\n <strong>Lane 1:</strong> Free DNA (100 ng).\n <strong>Lane 2:</strong> Free DNA incubated with 2U DpnII\n for 1 hr at 37°C (100 ng). <strong>Lane 3:</strong> Free DNA\n incubated with 2U MfeI for 1 hr at 37°C (100 ng). Migration\n patterns of DNA molecular weight markers are indicated.\n </span>\n </p>\n </div>\n </div>\n <aside class=\"image-picker__right\">\n <div class=\"image-picker__gallery\">\n <img\n alt=\"18-4101-dna-gel-data\"\n src=\"/content/images/products/nucleosomes/18-4101-dna-gel-data.png\"\n width=\"200\"\n class=\"image-picker__side-image image-picker__side-image_active\"\n role=\"button\"\n />\n </div>\n </aside>\n </section>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Technical Information</h3>\n <div\n class=\"ProductDescriptionContainer product-droppdown__section-description\"\n >\n <div class=\"product-tech-info\">\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Formulation</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n 50 µg lyophilized ST601-GATC1 DNA.\n </div>\n </div>\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Storage and Stability</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n Stable for 2 years at -20°C from date of receipt. After\n resuspending, aliquots should be stored at -80°C.\n </div>\n </div>\n </div>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Application Notes</h3>\n <div\n class=\"ProductDescriptionContainer product-droppdown__section-description\"\n >\n <p>\n ST601-GATC1 DNA is useful as a positive control for restriction\n enzyme accessibility nucleosome remodeling assays using the\n Biotinylated EpiDyne Remodeling Assay Substrate, as a DpnII (GATC in\n red, see DNA Sequence) restriction enzyme site are present within the 601 sequence.\n The naturally occurring MfeI restriction site (AATTGG in bold, see DNA Sequence)\n remains present as well.\n </p>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">DNA Sequence</h3>\n <div\n class=\"ProductDescriptionContainer product-droppdown__section-description\">\n <p style=\"line-break: anywhere\">\n 5'-GAATTCATCAGAATCCCGGTGCCGAGGCC<strong><FONT COLOR=\"#ff0000\">GATC</FONT COLOR=\"#ff0000\"></strong><strong>AATTGG</strong>TCGTAGACAGCTCTAGCACCGCTTAAACGCACGTACGCGCTGTCCCCCGCGTTTTAACCGCCAAGGGGATTACTCCCTAGTCTCCAGGCACGTGTCAGATATATACATCGATGATGATGGATAGATGGATGATGGATGGATGGATGATGATGGATGAATAGATGGATGGATGAAGCTT-3’\n </p>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">References</h3>\n <div\n class=\"ProductDescriptionContainer product-droppdown__section-description\"\n >\n <strong>Background References:</strong>\n <br />\n [1] Lowary & Widom <em>J. Mol. Biol.</em> (1998). PMID:\n <a\n href=\"https://pubmed.ncbi.nlm.nih.gov/9514715/\"\n target=\"_blank\"\n title=\"New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning\"\n >9514715</a\n ><br />\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Documents & Resources</h3>\n <div\n class=\"ProductDescriptionContainer product-droppdown__section-description\"\n >\n <div class=\"product-documents\">\n <a\n href=\"/content/documents/tds/18-4101.pdf\"\n target=\"_blank\"\n class=\"product-documents__link\"\n >\n <svg\n version=\"1.1\"\n id=\"Layer_1\"\n xmlns=\"http://www.w3.org/2000/svg\"\n xmlns:xlink=\"http://www.w3.org/1999/xlink\"\n x=\"0px\"\n y=\"0px\"\n viewBox=\"0 0 228 240\"\n style=\"enable-background: new 0 0 228 240\"\n xml:space=\"preserve\"\n class=\"product-documents__icon\"\n alt=\"16-0030 Datasheet\"\n >\n <g>\n <path\n class=\"product-documents__svg-pdf\"\n d=\"M191.92,68.77l-47.69-47.69c-1.33-1.33-3.12-2.08-5.01-2.08H45.09C41.17,19,38,22.17,38,26.09v184.36\n c0,3.92,3.17,7.09,7.09,7.09h141.82c3.92,0,7.09-3.17,7.09-7.09V73.8C194,71.92,193.25,70.1,191.92,68.77z M177.65,77.06h-41.7\n v-41.7L177.65,77.06z M178.05,201.59H53.95V34.95h66.92v47.86c0,5.14,4.17,9.31,9.31,9.31h47.86V201.59z\"\n />\n </g>\n <rect\n x=\"20\"\n y=\"112\"\n class=\"product-documents__svg-background\"\n width=\"146\"\n height=\"76\"\n />\n <g>\n <path\n class=\"product-documents__svg-pdf\"\n d=\"M23.83,125.68h22.36c5.29,0,9.41,1.33,12.35,4c2.94,2.67,4.42,6.39,4.42,11.18c0,4.78-1.47,8.51-4.42,11.18\n c-2.94,2.67-7.06,4-12.35,4H34.59v18.29H23.83V125.68z M44.81,147.9c5.38,0,8.07-2.32,8.07-6.97c0-2.39-0.67-4.16-2-5.31\n c-1.33-1.15-3.36-1.73-6.07-1.73H34.59v14.01H44.81z\"\n />\n <path\n class=\"product-documents__svg-pdf\"\n d=\"M69.92,125.68h18.91c5.29,0,9.84,0.97,13.66,2.9c3.82,1.93,6.74,4.72,8.76,8.35\n c2.02,3.63,3.04,7.98,3.04,13.04c0,5.06-1,9.42-3,13.08c-2,3.66-4.91,6.45-8.73,8.38c-3.82,1.93-8.4,2.9-13.73,2.9H69.92V125.68z\n M88.07,165.63c10.35,0,15.52-5.22,15.52-15.66c0-10.4-5.17-15.59-15.52-15.59h-7.38v31.26H88.07z\"\n />\n <path\n class=\"product-documents__svg-pdf\"\n d=\"M122.57,125.68h32.84v8.49h-22.22v11.18h20.84v8.49h-20.84v20.49h-10.63V125.68z\"\n />\n </g>\n </svg>\n <span class=\"product-documents__info\">Technical Datasheet</span>\n </a>\n </div>\n </div>\n </div>\n </div>\n </div>\n</div>\n","tags":[],"warranty":"","price":{"without_tax":{"formatted":"$115.00","value":115,"currency":"USD"},"tax_label":"Sales Tax"},"detail_messages":"","availability":"","page_title":"EpiDyne Remodeling Assay Substrate DNA (Positive Control)","cart_url":"https://www.epicypher.com/cart.php","max_purchase_quantity":0,"mpn":null,"upc":null,"shipping_messages":[],"rating":0,"meta_keywords":"ATPase, EpiDyne Nucleosome Remodeling Assay Substrate ST601-GATC1, EpiDyne, chromatin remodeling, chromatin remodeler, swi.snf, swi, snf, chrac, rsc, SMARCA, BRM1, BRG1, 16-4101, nucleosome remodeling, nucleosome sliding, sliding, restriction enzyme assay, DAM methyltransferase assay","show_quantity_input":1,"title":"EpiDyne® Remodeling Assay Substrate DNA ST601-GATC1","gift_wrapping_available":false,"min_purchase_quantity":0,"customizations":[],"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/374/292/DNA__93069.1614637188.gif?c=2","alt":"EpiDyne® Remodeling Assay Substrate DNA ST601-GATC1"}]} Pack Size: 50 µg
Description
EpiDyne® Remodeling Assay Substrate DNA ST601-GATC1 is a 217 base-pair double-stranded DNA fragment containing the 601 nucleosome positioning sequence [1], which has high affinity for histone octamers and is useful for nucleosome assembly. The DNA also includes a 3’ acceptor sequence to accommodate the histone octamer subsequent to remodeling. This positive control has a DpnII restriction site within the 601 sequence. When paired with the negative control DNA (EpiCypher 18-4100) these controls illustrate the migration range for the Restriction Enzyme Assay. See the EpiDyne Nucleosome Remodeling Assay Tech Note (Restriction Enzyme Assay) for more information.
Validation Data
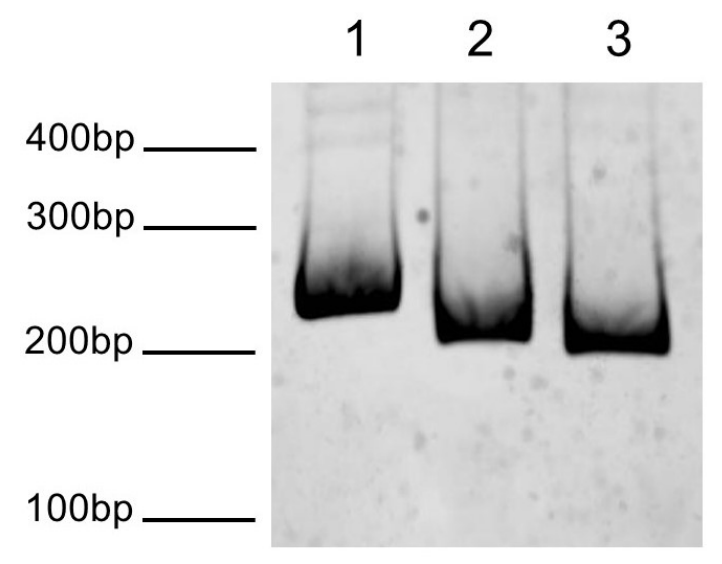
Figure 1: DNA gel data
ST601-GATC1 DNA resolved via native PAGE gel and stained
with ethidium bromide to visualize DNA.
Lane 1: Free DNA (100 ng).
Lane 2: Free DNA incubated with 2U DpnII
for 1 hr at 37°C (100 ng). Lane 3: Free DNA
incubated with 2U MfeI for 1 hr at 37°C (100 ng). Migration
patterns of DNA molecular weight markers are indicated.
Technical Information
Application Notes
ST601-GATC1 DNA is useful as a positive control for restriction enzyme accessibility nucleosome remodeling assays using the Biotinylated EpiDyne Remodeling Assay Substrate, as a DpnII (GATC in red, see DNA Sequence) restriction enzyme site are present within the 601 sequence. The naturally occurring MfeI restriction site (AATTGG in bold, see DNA Sequence) remains present as well.
DNA Sequence
5'-GAATTCATCAGAATCCCGGTGCCGAGGCCGATCAATTGGTCGTAGACAGCTCTAGCACCGCTTAAACGCACGTACGCGCTGTCCCCCGCGTTTTAACCGCCAAGGGGATTACTCCCTAGTCTCCAGGCACGTGTCAGATATATACATCGATGATGATGGATAGATGGATGATGGATGGATGGATGATGATGGATGAATAGATGGATGGATGAAGCTT-3’