The best histone PTM antibodies. Period.
EpiCypher takes a unique approach to validating histone post-translational modification (PTM) antibodies. Our exclusive SNAP Spike-in technology is the only method that uses physiological nucleosome controls to directly quantify antibody performance in ChIP, CUT&RUN, and CUT&Tag workflows. The resulting SNAP-Certified™ Antibodies have multiple advantages over competitors:
- Superior target specificity and affinity
- Robust performance in CUT&RUN and CUT&Tag
- Improved PTM profiling from low cell numbers
- Increased reliability through lot-specific testing
Have Questions?
We’re here to help. Click below and a member of our team will get back to you shortly!
Request More InfoIf you can’t trust your antibody, you can’t trust your data
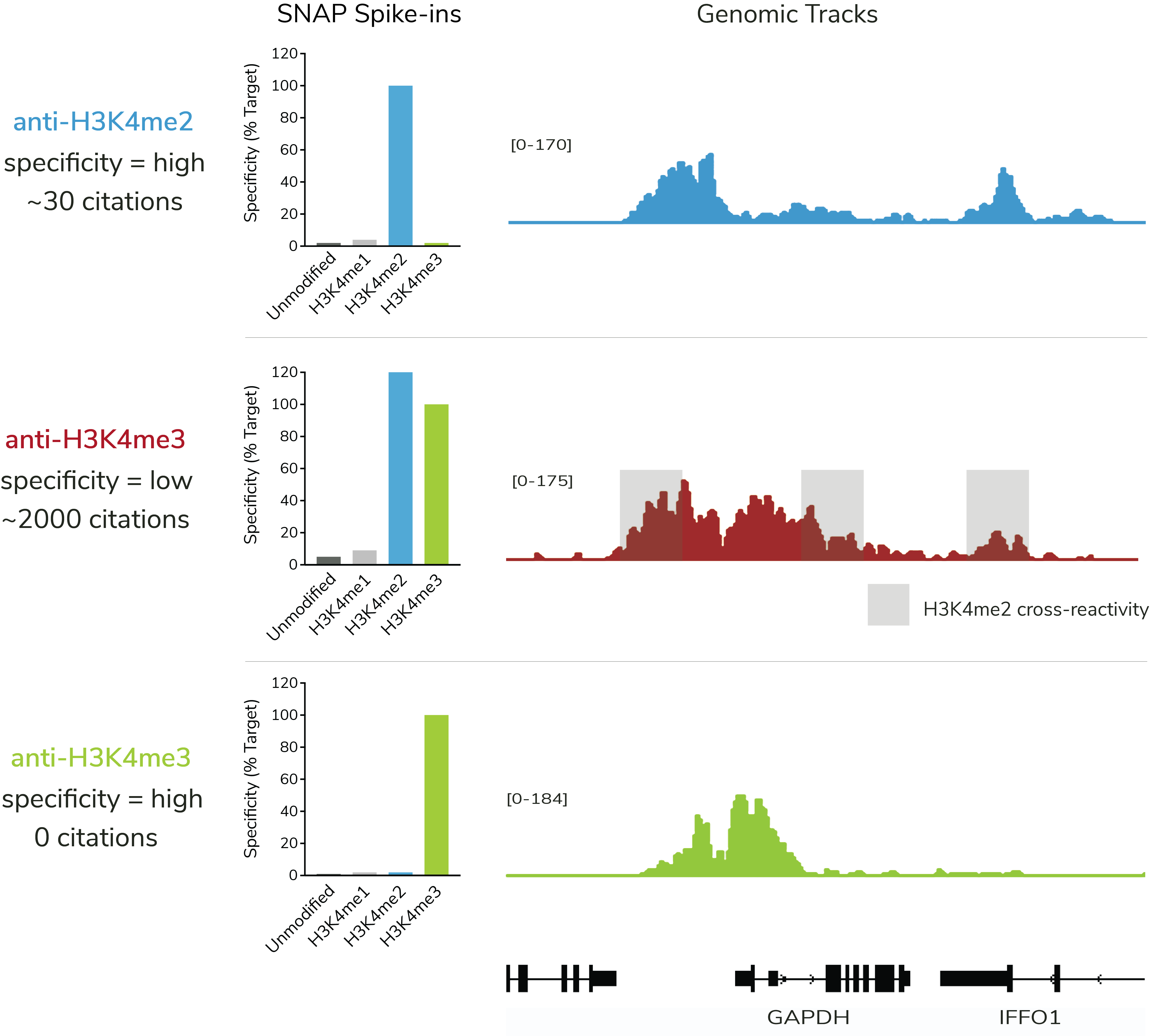
Work by EpiCypher and others show that over 70% of ChIP antibodies to histone PTMs have unacceptable rates of cross-reactivity and poor target affinity. These results challenge our understanding of PTM biology and demand new tools for improved chromatin mapping.
SNAP Spike-in Certification is the only method that directly quantifies histone PTM antibody specificity and binding efficiency in genomic mapping assays (see Figure).
Don’t question your results – EpiCypher offers the best PTM antibodies.
Robust validation metrics for exceptional antibodies
The only fit-for-purpose antibodies for epigenomics
Specificity is Crucial
Accurate biological interpretations from CUT&RUN and CUT&Tag assays require specific antibodies.
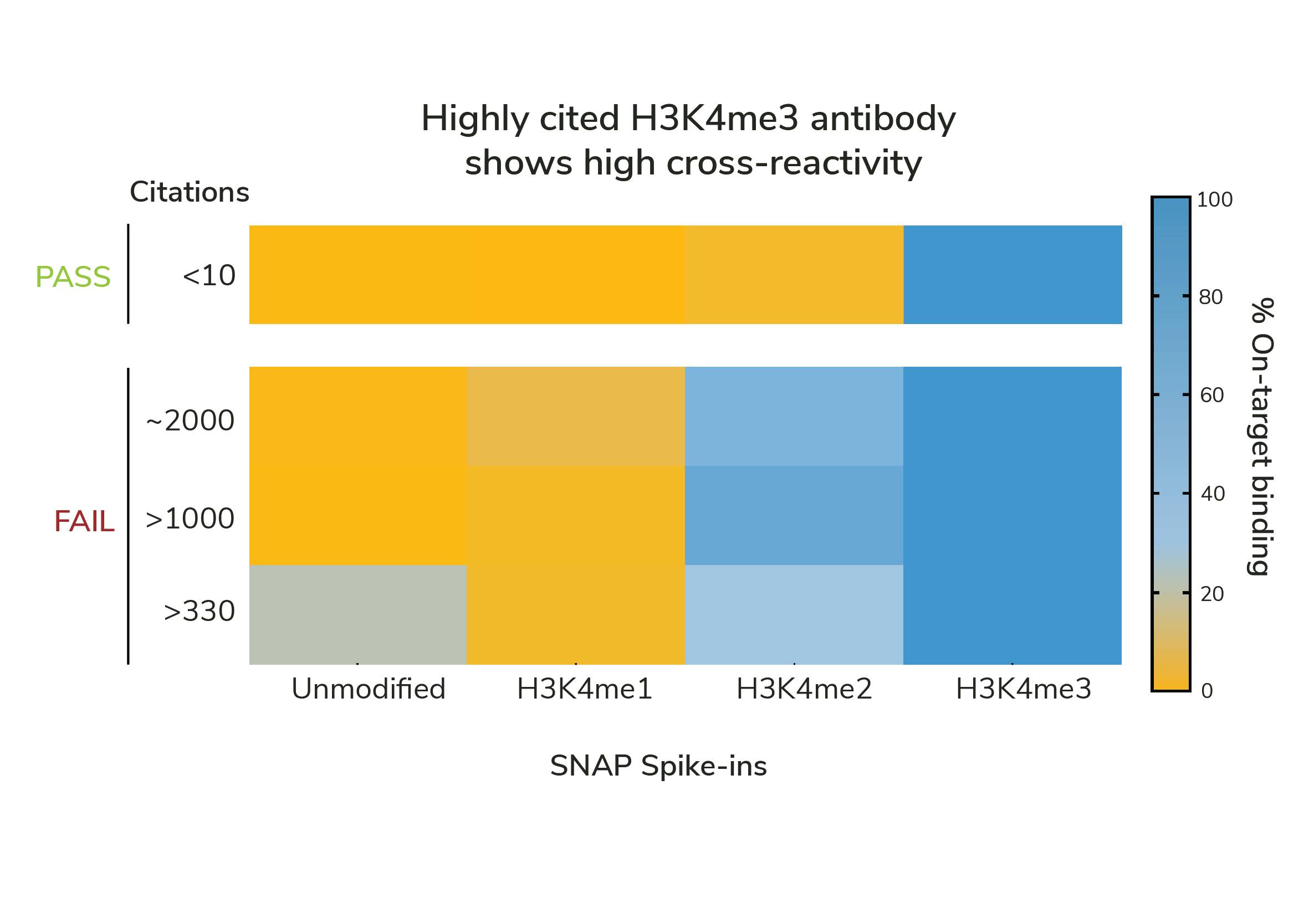
Standard validation methods cannot distinguish specific vs. nonspecific PTM antibodies. Even highly cited antibodies fail to meet minimal expectations for on-target profiling (see Figure).
SNAP-Certified Antibodies exhibit less than 20% cross-reactivity, ensuring reliable data with exquisite accuracy.
Efficiency Matters
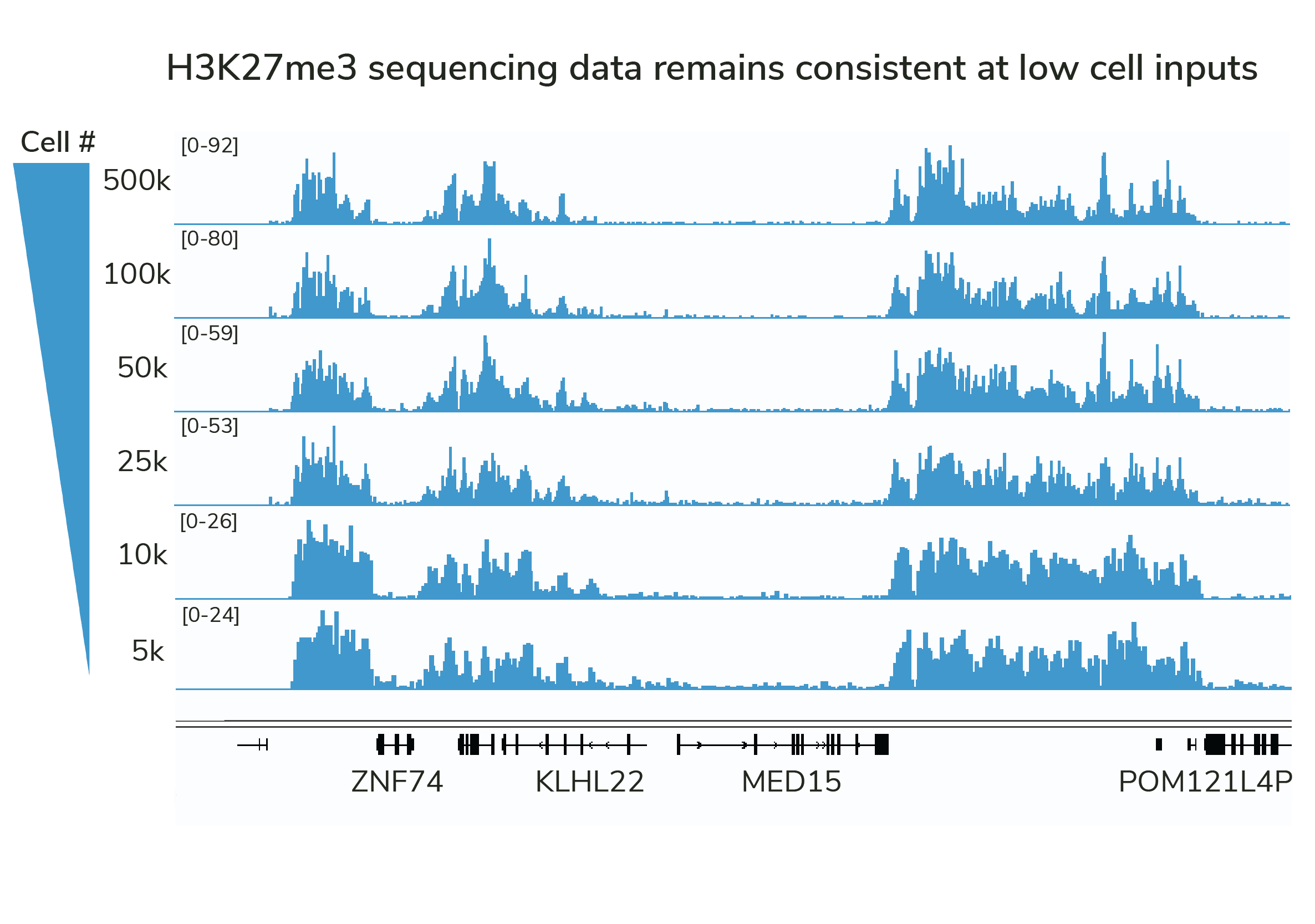
Highly efficient CUT&Tag and CUT&RUN antibodies enable reliable mapping from reduced cell numbers.
However, the historical focus on PTM specificity means that most antibodies lack the efficiency required for low- or single-cell mapping.
SNAP-Certified Antibodies are validated for high efficiency and generate reproducible profiles in cell titrations (see Figure).
Interested in SNAP-ChIP Certified Antibodies?
SNAP-CUTANA™ Spike-in Controls
Defined controls for reliable CUT&RUN and CUT&Tag assays. Panel available for histone lysine methylation PTMs.
SNAP-Certified CUT&RUN Antibodies
Highly specific and efficient PTM antibodies validated in CUT&RUN workflows using SNAP-CUTANA Spike-in Controls.
SNAP-ChIP® Certified Antibodies
ChIP antibodies validated using SNAP-ChIP Spike-ins. Display best-in-class specificity and target enrichment.