HA Tag CUTANA™ CUT&RUN Antibody
{"url":"https://www.epicypher.com/products/antibodies/cutana-cut-run-compatible-antibodies/ha-tag-cutana-cut-and-run-antibody","add_this":[{"service":"facebook","annotation":""},{"service":"email","annotation":""},{"service":"print","annotation":""},{"service":"twitter","annotation":""},{"service":"linkedin","annotation":""}],"gtin":"","id":"875","bulk_discount_rates":[],"can_purchase":true,"meta_description":"Rabbit polyclonal HA Tag antibody validated for CUT&RUN and WB. Rigorously tested for reliable and robust performance in CUT&RUN assays.","category":["Antibodies/CUTANA™ CUT&RUN Antibodies","Antibodies/CUTANA™ CUT&RUN Antibodies/CUT&RUN Antibodies - Chromatin-Associated Proteins","Epigenetics Kits and Reagents/CUTANA™ ChIC / CUT&RUN Assays"],"AddThisServiceButtonMeta":"","main_image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/875/937/ha-tag-cutana-cutandrun-antibody__43540.1645734529.jpg?c=2","alt":"HA Tag CUTANA CUTandRUN Antibody"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=875","shipping":{"calculated":true},"num_reviews":0,"weight":"0.00 LBS","custom_fields":[{"id":"909","name":"Pack Size","value":"100 µg"}],"sku":"13-2010","description":"<div class=\"product-general-info\">\n <ul class=\"product-general-info__list-left\">\n <li class=\"product-general-info__list-item\">\n <strong>Type: </strong>Polyclonal\n </li>\n <li class=\"product-general-info__list-item\">\n <strong>Host: </strong>Rabbit\n </li>\n <li class=\"product-general-info__list-item\">\n <strong>Applications: </strong>CUT&RUN, WB\n </li>\n </ul>\n <ul class=\"product-general-info__list-right\">\n <li class=\"product-general-info__list-item\">\n <strong>Reactivity: </strong>HA Epitope (YPYDVPDYA)\n </li>\n <li class=\"product-general-info__list-item\">\n <strong>Format: </strong>Antigen affinity-purified\n </li>\n <li class=\"product-general-info__list-item\">\n <strong>Target Size: </strong>N/A\n </li>\n </ul>\n <ul style=\"padding: 0\" class=\"product-general-info__list-right\">\n <li class=\"product-general-info__list-item\">\n <a href=\"#bioz\">\n <div\n id=\"w-s-3835-13-2010\"\n style=\"\n width: 240px;\n height: 58px;\n position: relative;\n overflow-y: hidden;\n \"\n ></div>\n <div id=\"bioz-w-pb-13-2010-div\" style=\"width: 240px\">\n <a\n id=\"bioz-w-pb-13-2010\"\n style=\"font-size: 12px; color: transparent\"\n href=\"https://www.bioz.com/\"\n target=\"_blank\"\n >\n <img\n src=\"https://cdn.bioz.com/assets/favicon.png\"\n style=\"\n width: 11px;\n height: 11px;\n vertical-align: baseline;\n padding-bottom: 0px;\n margin-left: 0px;\n margin-bottom: 0px;\n float: none;\n display: none;\n \"\n />\n Powered by Bioz\n </a>\n </div>\n </a>\n </li>\n </ul>\n</div>\n<div class=\"service_accordion product-droppdown\">\n <div class=\"container\">\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel current\">\n <h3 class=\"sub-title1\">Description</h3>\n <div\n class=\"ProductDescriptionContainer product-droppdown__section-description-specific\"\n >\n <p>\n This antibody meets EpiCypher’s \"CUTANA Compatible\" criteria\n for performance in Cleavage Under Targets and Release Using\n Nuclease (CUT&RUN) and/or Cleavage Under Targets and\n Tagmentation (CUT&Tag) approaches to genomic mapping. Every\n lot of a CUTANA Compatible antibody is tested in the indicated\n approach using\n <a href=\"/protocols\">EpiCypher optimized protocols</a> and\n determined to yield peaks that show a genomic distribution\n pattern consistent with reported function(s) of the target. HA\n antibody is useful for studies utilizing HA-tagged target\n proteins. HA Tag antibody produces CUT&RUN peaks (<strong\n >Figure 1</strong\n >) that overlap with GATA3 DNA-binding motifs (<strong\n >Figure 2</strong\n >) in breast cancer cells expressing 3xHA-tagged GATA3\n transcription factor [<a\n href=\"https://pubmed.ncbi.nlm.nih.gov/26922637/\"\n >Takaku et al., 2016</a\n >]*\n </p>\n <p style=\"\n background-color: #4698cb;\n color: #fff;\n padding: 1.3rem;\n text-align: center;\n border-radius: 12px;\n \">\n <a target=\"_blank\" style=\"color: #fff;\" href=\"/products/nucleosomes/snap-cutana-ha-tag-panel\">Pair with our SNAP-CUTANA™ HA Tag Panel for CUT&RUN success. Check it out here.\n </a>\n </p>\n <i>\n *Thanks to Dr. Takaku (UND) for 3xFlag-GATA3-3xHA MDA-MB-231\n cells.\n </i>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel current\">\n <h3 class=\"sub-title1\">Validation Data</h3>\n <div\n class=\"ProductDescriptionContainer product-droppdown__section-description-specific\"\n >\n <section class=\"image-picker\">\n <div class=\"image-picker__left\">\n <div\n class=\"image-picker__main-content_active image-picker__main-content\"\n >\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg\n class=\"image-picker__svg-left\"\n width=\"24\"\n height=\"24\"\n viewBox=\"0 0 24 24\"\n >\n <path\n d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\"\n />\n </svg>\n </button>\n <a\n href=\"/content/images/products/antibodies/13-2010-HA-Tag-Heatmap.jpeg\"\n target=\"_new\"\n class=\"image-picker__main-image-link\"\n ><img\n loading=\"lazy\"\n alt=\"13-2010-HA-Tag-Heatmap\"\n src=\"/content/images/products/antibodies/13-2010-HA-Tag-Heatmap.jpeg\"\n class=\"image-picker__main-image\"\n />\n <span class=\"image-picker__main-image-caption\"\n >(Click to enlarge)</span\n ></a\n >\n <button class=\"image-picker__right-arrow\">\n <svg\n class=\"image-picker__svg-right\"\n width=\"24\"\n height=\"24\"\n viewBox=\"0 0 24 24\"\n >\n <path\n d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\"\n />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\"\n ><strong>Figure 1: HA Tag peaks in CUT&RUN </strong\n ><br />\n CUT&RUN was performed as described in\n <strong>Figure 5</strong>. Peaks were called using\n MACS2. (<strong>A</strong>) Heatmaps show GATA3-3xHA\n peaks relative to IgG and H3K4me3 control antibodies\n in aligned rows ranked by intensity (top to bottom)\n and colored such that red indicates high localized\n enrichment and blue denotes background signal.\n (<strong>B</strong>) The number of GATA3-3xHA peaks\n that fall into distinct classes of functionally\n annotated genomic regions are shown.\n </span>\n </p>\n </div>\n <div class=\"image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg\n class=\"image-picker__svg-left\"\n width=\"24\"\n height=\"24\"\n viewBox=\"0 0 24 24\"\n >\n <path\n d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\"\n />\n </svg>\n </button>\n <a\n href=\"/content/images/products/antibodies/13-2010-HA-Tag-Motif.jpeg\"\n target=\"_new\"\n class=\"image-picker__main-image-link\"\n ><img\n loading=\"lazy\"\n alt=\"13-2010-HA-Tag-Motif\"\n src=\"/content/images/products/antibodies/13-2010-HA-Tag-Motif.jpeg\"\n class=\"image-picker__main-image\"\n />\n <span class=\"image-picker__main-image-caption\"\n >(Click to enlarge)</span\n ></a\n >\n <button class=\"image-picker__right-arrow\">\n <svg\n class=\"image-picker__svg-right\"\n width=\"24\"\n height=\"24\"\n viewBox=\"0 0 24 24\"\n >\n <path\n d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\"\n />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\"\n ><strong\n >Figure 2: HA-tagged transcription factor binding\n motif analysis in CUT&RUN</strong\n ><br />\n (<strong>A</strong>) Homer analysis determined that\n the GATA3 consensus motif, represented as a sequence\n logo position weight matrix, was enriched under\n GATA3-3xHA CUT&RUN peaks. (<strong>B</strong>) The\n number of GATA3-3xHA peaks containing GATA3 consensus\n motifs from panel A is represented by a Venn Diagram.\n (<strong>C-D</strong>) Two representative loci show\n overlap of GATA3-3xHA peaks with the consensus motifs\n noted by tick marks beneath the tracks.\n </span>\n </p>\n </div>\n\n <div class=\"image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg\n class=\"image-picker__svg-left\"\n width=\"24\"\n height=\"24\"\n viewBox=\"0 0 24 24\"\n >\n <path\n d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\"\n />\n </svg>\n </button>\n <a\n href=\"/content/images/products/antibodies/13-2010-western-blot.jpeg\"\n target=\"_new\"\n class=\"image-picker__main-image-link\"\n ><img\n loading=\"lazy\"\n alt=\"13-2010-western-blot\"\n src=\"/content/images/products/antibodies/13-2010-western-blot.jpeg\"\n class=\"image-picker__main-image\"\n />\n <span class=\"image-picker__main-image-caption\"\n >(Click to enlarge)</span\n ></a\n >\n <button class=\"image-picker__right-arrow\">\n <svg\n class=\"image-picker__svg-right\"\n width=\"24\"\n height=\"24\"\n viewBox=\"0 0 24 24\"\n >\n <path\n d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\"\n />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\"\n ><strong>Figure 3: Western blot data</strong><br />\n <em>E. coli</em> cells expressing a multi-tag fusion\n protein were used to prepare whole cell lysates. The\n indicated amounts (ng) of lysate were loaded onto a\n 4-20% SDS-PAGE gel and analyzed under standard western\n blot conditions using HA Tag antibody at a dilution of\n 1:25,000.\n </span>\n </p>\n </div>\n <div class=\"image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg\n class=\"image-picker__svg-left\"\n width=\"24\"\n height=\"24\"\n viewBox=\"0 0 24 24\"\n >\n <path\n d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\"\n />\n </svg>\n </button>\n <a\n href=\"/content/images/products/antibodies/13-2010-HA-Tag-ChAP-Spike-ins.jpeg\"\n target=\"_new\"\n class=\"image-picker__main-image-link\"\n ><img\n loading=\"lazy\"\n alt=\"13-2010-HA-Tag-ChAP-Spike-ins\"\n src=\"/content/images/products/antibodies/13-2010-HA-Tag-ChAP-Spike-ins.jpeg\"\n class=\"image-picker__main-image\"\n />\n <span class=\"image-picker__main-image-caption\"\n >(Click to enlarge)</span\n ></a\n >\n <button class=\"image-picker__right-arrow\">\n <svg\n class=\"image-picker__svg-right\"\n width=\"24\"\n height=\"24\"\n viewBox=\"0 0 24 24\"\n >\n <path\n d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\"\n />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\"\n ><strong\n >Figure 4: Target-specific epitope cleavage of HA\n Tag antibody in CUT&RUN was determined using\n DNA-barcoded recombinant nucleosome spike-in\n controls</strong\n ><br />\n (<strong>A</strong>) A panel of recombinant\n nucleosomes was created where various epitope tags\n (3xTY1, 3xFLAG, 3xHA) were fused to the histone H3\n tail. The fused nucleosomes and an unmodified control\n were immobilized to streptavidin beads (SA Bead) and\n spiked into CUT&RUN samples alongside ConA bead\n immobilized MDA-MB-231 cells expressing GATA3-3xHA\n (Figure 1). HA Tag antibody and pAG-MNase (EpiCypher\n <a\n href=\"https://www.epicypher.com/products/epigenetics-reagents-and-assays/cutana-pag-mnase-for-chic-cut-and-run-workflows\"\n >15-1016</a\n >) were then added to release antibody-bound\n nucleosomes into solution through pAG-MNase mediated\n cleavage of the linker DNA (light blue). This approach\n provided a defined experimental control to assess\n whether the HA Tag antibody selectively cleaved the\n target epitope with high specificity and minimal\n background. (<strong>B</strong>) CUT&RUN sequence\n reads were aligned to the unique DNA \"barcodes\"\n corresponding to each nucleosome in the spike-in\n panel. Data are expressed as the percent of reads\n recovered relative to the intended target (3xHA, set\n to 100%). This analysis confirms that the HA Tag\n antibody specifically liberated the target\n epitope-tagged nucleosome into solution.\n </span>\n </p>\n </div>\n <div class=\"image-picker__main-content\">\n <div class=\"image-picker__header-content\">\n <button class=\"image-picker__left-arrow\">\n <svg\n class=\"image-picker__svg-left\"\n width=\"24\"\n height=\"24\"\n viewBox=\"0 0 24 24\"\n >\n <path\n d=\"M16.67 0l2.83 2.829-9.339 9.175 9.339 9.167-2.83 2.829-12.17-11.996z\"\n />\n </svg>\n </button>\n <a\n href=\"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/original/image-manager/2022.07.06-c-r-workflow-simple-rgb-ew.png?t=1697220982&_gl=1*u66xe1*_ga*NDc4MDE2MzAxLjE2OTY2MjA3NTU.*_ga_WS2VZYPC6G*MTY5NzY5NzkxNi4zNi4xLjE2OTc2OTk4OTEuNTYuMC4w\"\n target=\"_new\"\n class=\"image-picker__main-image-link\"\n ><img\n loading=\"lazy\"\n alt=\"13_2012_IP\"\n src=\"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/original/image-manager/2022.07.06-c-r-workflow-simple-rgb-ew.png?t=1697220982&_gl=1*u66xe1*_ga*NDc4MDE2MzAxLjE2OTY2MjA3NTU.*_ga_WS2VZYPC6G*MTY5NzY5NzkxNi4zNi4xLjE2OTc2OTk4OTEuNTYuMC4w\"\n class=\"image-picker__main-image\"\n />\n <span class=\"image-picker__main-image-caption\"\n >(Click to enlarge)</span\n ></a\n >\n <button class=\"image-picker__right-arrow\">\n <svg\n class=\"image-picker__svg-right\"\n width=\"24\"\n height=\"24\"\n viewBox=\"0 0 24 24\"\n >\n <path\n d=\"M7.33 24l-2.83-2.829 9.339-9.175-9.339-9.167 2.83-2.829 12.17 11.996z\"\n />\n </svg>\n </button>\n </div>\n <p>\n <span class=\"image-picker__span-content\"\n ><strong>Figure 5: CUT&RUN Methods</strong><br />\n CUT&RUN was performed on 500k MDA-MB-231 cells stably\n expressing c-terminal 3xHA-tagged GATA3 [1]* with 0.5\n µg of either HA Tag, H3K4me3 positive control\n (EpiCypher\n <a\n href=\"/products/antibodies/snap-chip-certified-antibodies/histone-h3k4me3-antibody-snap-chip-certified-cutana-cut-run-compatible\"\n >13-0041</a\n >), or IgG negative control (EpiCypher\n <a\n href=\"/products/nucleosomes/snap-cutana-spike-in-controls/cutana-rabbit-igg-cut-run-negative-control-antibody\"\n >13-0042</a\n >) antibodies using the CUTANA™ ChIC/CUT&RUN Kit v2.0\n (EpiCypher\n <a\n href=\"/products/epigenetics-reagents-and-assays/cutana-chic-cut-and-run-kit\"\n >14-1048</a\n >). Library preparation was performed with 5 ng of DNA\n (or the total amount recovered if less than 5 ng)\n using the CUTANA™ CUT&RUN Library Prep Kit (EpiCypher\n <a\n href=\"/products/epigenetics-reagents-and-assays/cutana-cut-and-run-library-prep-kit\"\n >14-1001/14-1002</a\n >). Both kit protocols were adapted for high\n throughput Tecan liquid handling. Libraries were run\n on an Illumina NextSeq2000 with paired-end sequencing\n (2x50 bp). Sample sequencing depth was 2.4 million\n reads (IgG), 4.1 million reads (H3K4me3), and 8.6\n million reads (HA Tag). Data were aligned to the hg19\n genome using Bowtie2. Data were filtered to remove\n duplicates, multi-aligned reads, and ENCODE DAC Exclusion List\n regions.\n </span>\n </p>\n </div>\n </div>\n <aside class=\"image-picker__right\">\n <div class=\"image-picker__gallery\">\n <img\n loading=\"lazy\"\n alt=\"13-2010-HA-Tag-Heatmap\"\n src=\"/content/images/products/antibodies/13-2010-HA-Tag-Heatmap.jpeg\"\n width=\"200\"\n class=\"image-picker__side-image image-picker__side-image_active\"\n role=\"button\"\n />\n <img\n loading=\"lazy\"\n alt=\"13-2010-HA-Tag-Motif\"\n src=\"/content/images/products/antibodies/13-2010-HA-Tag-Motif.jpeg\"\n class=\"image-picker__side-image\"\n role=\"button\"\n />\n <img\n loading=\"lazy\"\n alt=\"13-2010-western-blot\"\n src=\"/content/images/products/antibodies/13-2010-western-blot.jpeg\"\n class=\"image-picker__side-image\"\n role=\"button\"\n />\n <img\n loading=\"lazy\"\n alt=\"13-2010-HA-Tag-ChAP-Spike-ins\"\n src=\"/content/images/products/antibodies/13-2010-HA-Tag-ChAP-Spike-ins.jpeg\"\n class=\"image-picker__side-image\"\n role=\"button\"\n />\n <img\n loading=\"lazy\"\n alt=\"13-2010-HA-Tag-ChAP-Spike-ins\"\n src=\"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/original/image-manager/2022.07.06-c-r-workflow-simple-rgb-ew.png?t=1697220982&_gl=1*u66xe1*_ga*NDc4MDE2MzAxLjE2OTY2MjA3NTU.*_ga_WS2VZYPC6G*MTY5NzY5NzkxNi4zNi4xLjE2OTc2OTk4OTEuNTYuMC4w\"\n class=\"image-picker__side-image\"\n role=\"button\"\n />\n </div>\n </aside>\n </section>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Technical Information</h3>\n <div\n class=\"ProductDescriptionContainer product-droppdown__section-description\"\n >\n <div class=\"product-tech-info\">\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Immunogen</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n A synthetic HA peptide (sequence: YPYDVPDYA)\n </div>\n </div>\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Storage</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n Stable for 1 year at 4°C from date of receipt\n </div>\n </div>\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Formulation</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n Antigen affinity-purified antibody in phosphate buffered\n saline (PBS), 0.09% sodium azide\n </div>\n </div>\n </div>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Recommended Dilution</h3>\n <div\n class=\"ProductDescriptionContainer product-droppdown__section-description\"\n >\n <div class=\"product-tech-info\">\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>CUT&RUN:</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n 0.5 µg per reaction\n </div>\n </div>\n <div class=\"product-tech-info__line-item\">\n <div class=\"product-tech-info__line-item-left\">\n <b>Western Blot (WB):</b>\n </div>\n <div class=\"product-tech-info__line-item-right\">\n 1:1,000 - 1:30,000\n </div>\n </div>\n </div>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Documents & Resources</h3>\n <div\n class=\"ProductDescriptionContainer product-droppdown__section-description\"\n >\n <div class=\"product-documents\">\n <a\n href=\"/content/documents/tds/13-2010.pdf\"\n target=\"_new\"\n class=\"product-documents__link\"\n >\n <svg\n version=\"1.1\"\n id=\"Layer_1\"\n xmlns=\"http://www.w3.org/2000/svg\"\n xmlns:xlink=\"http://www.w3.org/1999/xlink\"\n x=\"0px\"\n y=\"0px\"\n viewBox=\"0 0 228 240\"\n style=\"enable-background: new 0 0 228 240\"\n xml:space=\"preserve\"\n class=\"product-documents__icon\"\n >\n <g>\n <path\n class=\"product-documents__svg-pdf\"\n d=\"M191.92,68.77l-47.69-47.69c-1.33-1.33-3.12-2.08-5.01-2.08H45.09C41.17,19,38,22.17,38,26.09v184.36\n c0,3.92,3.17,7.09,7.09,7.09h141.82c3.92,0,7.09-3.17,7.09-7.09V73.8C194,71.92,193.25,70.1,191.92,68.77z M177.65,77.06h-41.7\n v-41.7L177.65,77.06z M178.05,201.59H53.95V34.95h66.92v47.86c0,5.14,4.17,9.31,9.31,9.31h47.86V201.59z\"\n />\n </g>\n <rect\n x=\"20\"\n y=\"112\"\n class=\"product-documents__svg-background\"\n width=\"146\"\n height=\"76\"\n />\n <g>\n <path\n class=\"product-documents__svg-pdf\"\n d=\"M23.83,125.68h22.36c5.29,0,9.41,1.33,12.35,4c2.94,2.67,4.42,6.39,4.42,11.18c0,4.78-1.47,8.51-4.42,11.18\n c-2.94,2.67-7.06,4-12.35,4H34.59v18.29H23.83V125.68z M44.81,147.9c5.38,0,8.07-2.32,8.07-6.97c0-2.39-0.67-4.16-2-5.31\n c-1.33-1.15-3.36-1.73-6.07-1.73H34.59v14.01H44.81z\"\n />\n <path\n class=\"product-documents__svg-pdf\"\n d=\"M69.92,125.68h18.91c5.29,0,9.84,0.97,13.66,2.9c3.82,1.93,6.74,4.72,8.76,8.35\n c2.02,3.63,3.04,7.98,3.04,13.04c0,5.06-1,9.42-3,13.08c-2,3.66-4.91,6.45-8.73,8.38c-3.82,1.93-8.4,2.9-13.73,2.9H69.92V125.68z\n M88.07,165.63c10.35,0,15.52-5.22,15.52-15.66c0-10.4-5.17-15.59-15.52-15.59h-7.38v31.26H88.07z\"\n />\n <path\n class=\"product-documents__svg-pdf\"\n d=\"M122.57,125.68h32.84v8.49h-22.22v11.18h20.84v8.49h-20.84v20.49h-10.63V125.68z\"\n />\n </g>\n </svg>\n <span class=\"product-documents__info\"\n >Technical Datasheet</span\n >\n </a>\n </div>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel\">\n <h3 class=\"sub-title1\">Additional Info</h3>\n <div\n class=\"ProductDescriptionContainer product-droppdown__section-description\"\n >\n <p>\n This product is provided for commercial sale under license\n from Bethyl Laboratories, Inc.\n </p>\n </div>\n </div>\n </div>\n <div id=\"prodAccordion\">\n <div id=\"ProductDescription\" class=\"Block Panel current\">\n <h3\n id=\"bioz\"\n style=\"padding-bottom: 5px; margin-top: 1rem\"\n class=\"sub-title1\"\n >\n Product References\n </h3>\n\n <div\n class=\"ProductDescriptionContainer product-droppdown__section-description-specific\"\n >\n <object\n id=\"wobj-3835-13-2010-q\"\n type=\"text/html\"\n data=\"https://www.bioz.com/v_widget_6_0/3835/13-2010/\"\n style=\"width: 100%; height: 193px\"\n ></object>\n <div id=\"bioz-w-pb-3835-13-2010-q-div\" style=\"width: 100%\">\n <a\n id=\"bioz-w-pb-3835-13-2010-q\"\n style=\"\n font-size: 12px;\n text-decoration: none;\n color: #4698cb;\n \"\n href=\"https://www.bioz.com/\"\n target=\"_blank\"\n ><img\n src=\"https://cdn.bioz.com/assets/favicon.png\"\n style=\"\n width: 11px;\n height: 11px;\n vertical-align: baseline;\n padding-bottom: 0px;\n margin-left: 0px;\n margin-bottom: 0px;\n float: none;\n \"\n />\n Powered by Bioz</a\n >\n <a\n style=\"\n font-size: 12px;\n text-decoration: none;\n float: right;\n color: transparent;\n \"\n href=\"https://www.bioz.com/result/13-2010/product/EpiCypher/?cn=13-2010\"\n target=\"_blank\"\n >\n See more details on Bioz</a\n >\n </div>\n </div>\n </div>\n </div>\n </div>\n</div>\n<script>\n $(document).ready(function () {\n var widget_micro_obj = new v_widget_obj(\"s\", [1]);\n widget_micro_obj.request_catalog_number_widget_data_internal(\n \"13-2010\",\n \"13-2010\"\n );\n });\n</script>\n<style>\n .bioz-w-header td {\n padding: 0;\n }\n td table {\n margin: 0;\n padding: 6px !important;\n }\n</style>","tags":[],"warranty":"","price":{"without_tax":{"formatted":"$495.00","value":495,"currency":"USD"},"tax_label":"Sales Tax"},"detail_messages":"","availability":"","page_title":"HA Tag Antibody | CUTANA™ CUT&RUN Compatible","cart_url":"https://www.epicypher.com/cart.php","max_purchase_quantity":0,"mpn":"","upc":null,"options":[],"related_products":[{"id":772,"sku":"14-1048","name":"CUTANA™ ChIC/CUT&RUN Kit","url":"https://www.epicypher.com/products/epigenetics-reagents-and-assays/cutana-chic-cut-and-run-kit","availability":"","rating":null,"brand":{"name":null},"category":["Epigenetics Kits and Reagents","Epigenetics Kits and Reagents/CUTANA™ ChIC / CUT&RUN Assays"],"summary":"\n \n \n \n \n \n \n \n ","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/772/998/Epicypher_CUTANA_CUTRUN_Box__31908.1646406725.png?c=2","alt":"CUTANA™ ChIC/CUT&RUN Kit"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/772/998/Epicypher_CUTANA_CUTRUN_Box__31908.1646406725.png?c=2","alt":"CUTANA™ ChIC/CUT&RUN Kit"}],"date_added":"13th Aug 2020","pre_order":false,"show_cart_action":true,"has_options":false,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":[{"id":588,"name":"Pack size","value":"48 Reactions"}],"num_reviews":null,"weight":{"formatted":"0.01 LBS","value":0.01},"demo":false,"add_to_cart_url":"https://www.epicypher.com/cart.php?action=add&product_id=772","price":{"without_tax":{"currency":"USD","formatted":"$945.00","value":945},"tax_label":"Sales Tax"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=772"},{"id":889,"sku":null,"name":"CUTANA™ CUT&RUN Library Prep Kit","url":"https://www.epicypher.com/products/epigenetics-reagents-and-assays/cutana-cut-and-run-library-prep-kit","availability":"","rating":null,"brand":{"name":null},"category":["Epigenetics Kits and Reagents","Epigenetics Kits and Reagents/CUTANA™ ChIC / CUT&RUN Assays"],"summary":"\n \n \n \n \n \n \n \n ","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/889/997/Epicypher_CUTANA_CUTRUN_LibraryPrep_Box__64431.1646406521.png?c=2","alt":"CUTANA™ CUT&RUN Library Prep Kit"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/889/997/Epicypher_CUTANA_CUTRUN_LibraryPrep_Box__64431.1646406521.png?c=2","alt":"CUTANA™ CUT&RUN Library Prep Kit"}],"date_added":"31st Jan 2022","pre_order":false,"show_cart_action":true,"has_options":true,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":[{"id":983,"name":"Pack Size","value":"48 Reactions"}],"num_reviews":null,"weight":{"formatted":"0.01 LBS","value":0.01},"demo":false,"price":{"without_tax":{"currency":"USD","formatted":"$1,525.00","value":1525},"tax_label":"Sales Tax"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=889"},{"id":694,"sku":null,"name":"CUTANA™ pAG-MNase for ChIC/CUT&RUN Workflows","url":"https://www.epicypher.com/products/epigenetics-reagents-and-assays/cutana-pag-mnase-for-chic-cut-and-run-workflows","availability":"","rating":null,"brand":{"name":null},"category":["Epigenetics Kits and Reagents","Epigenetics Kits and Reagents/CUTANA™ ChIC / CUT&RUN Assays"],"summary":"\n \n \n Type: Nuclease\n \n \n Mol Wgt: 43.7 kDa\n \n \n \n ","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/694/689/Screen_Shot_2020-02-12_at_11.01.55_AM__17144.1581530752.png?c=2","alt":"CUTANA™ pAG-MNase for ChIC/CUT&RUN Workflows"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/694/689/Screen_Shot_2020-02-12_at_11.01.55_AM__17144.1581530752.png?c=2","alt":"CUTANA™ pAG-MNase for ChIC/CUT&RUN Workflows"}],"date_added":"12th Aug 2019","pre_order":false,"show_cart_action":true,"has_options":true,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":[{"id":1174,"name":"Internal Comment","value":"Excess in bottom of Venom"},{"id":1175,"name":"Internal Comment","value":"Bulk in Psylocke"}],"num_reviews":null,"weight":{"formatted":"0.01 LBS","value":0.01},"demo":false,"price":{"without_tax":{"currency":"USD","formatted":"$335.00","value":335},"price_range":{"min":{"without_tax":{"currency":"USD","formatted":"$335.00","value":335},"tax_label":"Sales Tax"},"max":{"without_tax":{"currency":"USD","formatted":"$1,295.00","value":1295},"tax_label":"Sales Tax"}},"tax_label":"Sales Tax"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=694"},{"id":758,"sku":"13-0042","name":"CUTANA™ IgG Negative Control Antibody for CUT&RUN and CUT&Tag","url":"https://www.epicypher.com/products/nucleosomes/snap-cutana-spike-in-controls/cutana-igg-negative-control-antibody-for-cut-run-and-cut-tag","availability":"","rating":null,"brand":{"name":null},"category":["Nucleosomes/SNAP-CUTANA™ Spike-in Controls","Antibodies/CUTANA™ CUT&RUN Antibodies","Epigenetics Kits and Reagents","Epigenetics Kits and Reagents/CUTANA™ ChIC / CUT&RUN Assays","Epigenetics Kits and Reagents/CUTANA™ CUT&Tag Assays"],"summary":"\n \n \n Type: Polyclonal\n \n \n Host: Rabbit\n \n \n Applications: CUT&R","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/758/739/1579731877.1280.1280__61771.1592491196.png?c=2","alt":"CUTANA™ IgG Negative Control Antibody for CUT&RUN and CUT&Tag"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/758/739/1579731877.1280.1280__61771.1592491196.png?c=2","alt":"CUTANA™ IgG Negative Control Antibody for CUT&RUN and CUT&Tag"}],"date_added":"17th Jun 2020","pre_order":false,"show_cart_action":true,"has_options":false,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":[{"id":1148,"name":"Pack Size","value":"100 µg"},{"id":1149,"name":"Internal Comment","value":"bulk stocks from Thermo in cold room"}],"num_reviews":null,"weight":{"formatted":"0.01 LBS","value":0.01},"demo":false,"add_to_cart_url":"https://www.epicypher.com/cart.php?action=add&product_id=758","price":{"without_tax":{"currency":"USD","formatted":"$105.00","value":105},"tax_label":"Sales Tax"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=758"}],"shipping_messages":[],"rating":0,"meta_keywords":"HA Tag antibody, epitope tag antibody, CUT&RUN antibody","show_quantity_input":1,"title":"HA Tag CUTANA™ CUT&RUN Antibody","gift_wrapping_available":false,"min_purchase_quantity":0,"customizations":[],"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/875/937/ha-tag-cutana-cutandrun-antibody__43540.1645734529.jpg?c=2","alt":"HA Tag CUTANA CUTandRUN Antibody"}]} Pack Size: 100 µg
- Type: Polyclonal
- Host: Rabbit
- Applications: CUT&RUN, WB
- Reactivity: HA Epitope (YPYDVPDYA)
- Format: Antigen affinity-purified
- Target Size: N/A
Description
This antibody meets EpiCypher’s "CUTANA Compatible" criteria for performance in Cleavage Under Targets and Release Using Nuclease (CUT&RUN) and/or Cleavage Under Targets and Tagmentation (CUT&Tag) approaches to genomic mapping. Every lot of a CUTANA Compatible antibody is tested in the indicated approach using EpiCypher optimized protocols and determined to yield peaks that show a genomic distribution pattern consistent with reported function(s) of the target. HA antibody is useful for studies utilizing HA-tagged target proteins. HA Tag antibody produces CUT&RUN peaks (Figure 1) that overlap with GATA3 DNA-binding motifs (Figure 2) in breast cancer cells expressing 3xHA-tagged GATA3 transcription factor [Takaku et al., 2016]*
Pair with our SNAP-CUTANA™ HA Tag Panel for CUT&RUN success. Check it out here.
*Thanks to Dr. Takaku (UND) for 3xFlag-GATA3-3xHA MDA-MB-231 cells.Validation Data
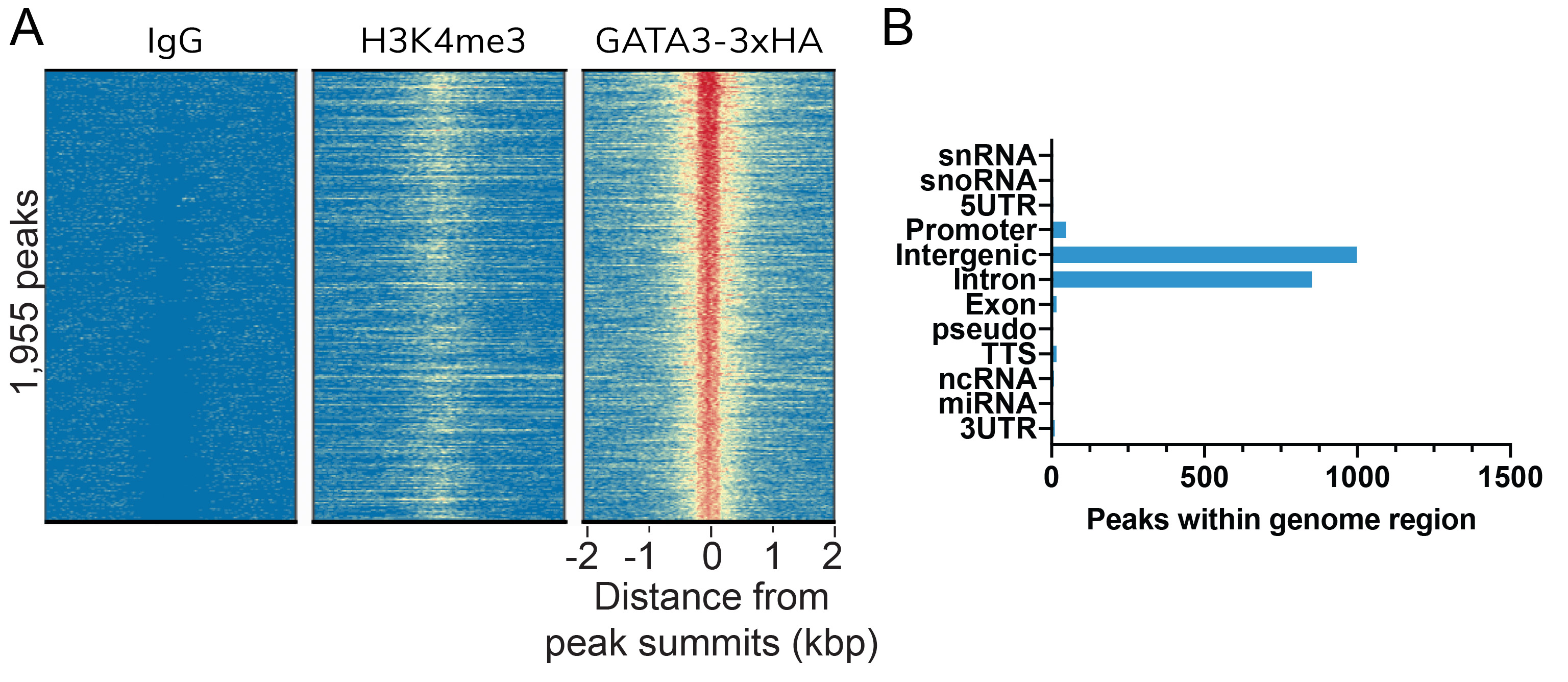
Figure 1: HA Tag peaks in CUT&RUN
CUT&RUN was performed as described in
Figure 5. Peaks were called using
MACS2. (A) Heatmaps show GATA3-3xHA
peaks relative to IgG and H3K4me3 control antibodies
in aligned rows ranked by intensity (top to bottom)
and colored such that red indicates high localized
enrichment and blue denotes background signal.
(B) The number of GATA3-3xHA peaks
that fall into distinct classes of functionally
annotated genomic regions are shown.
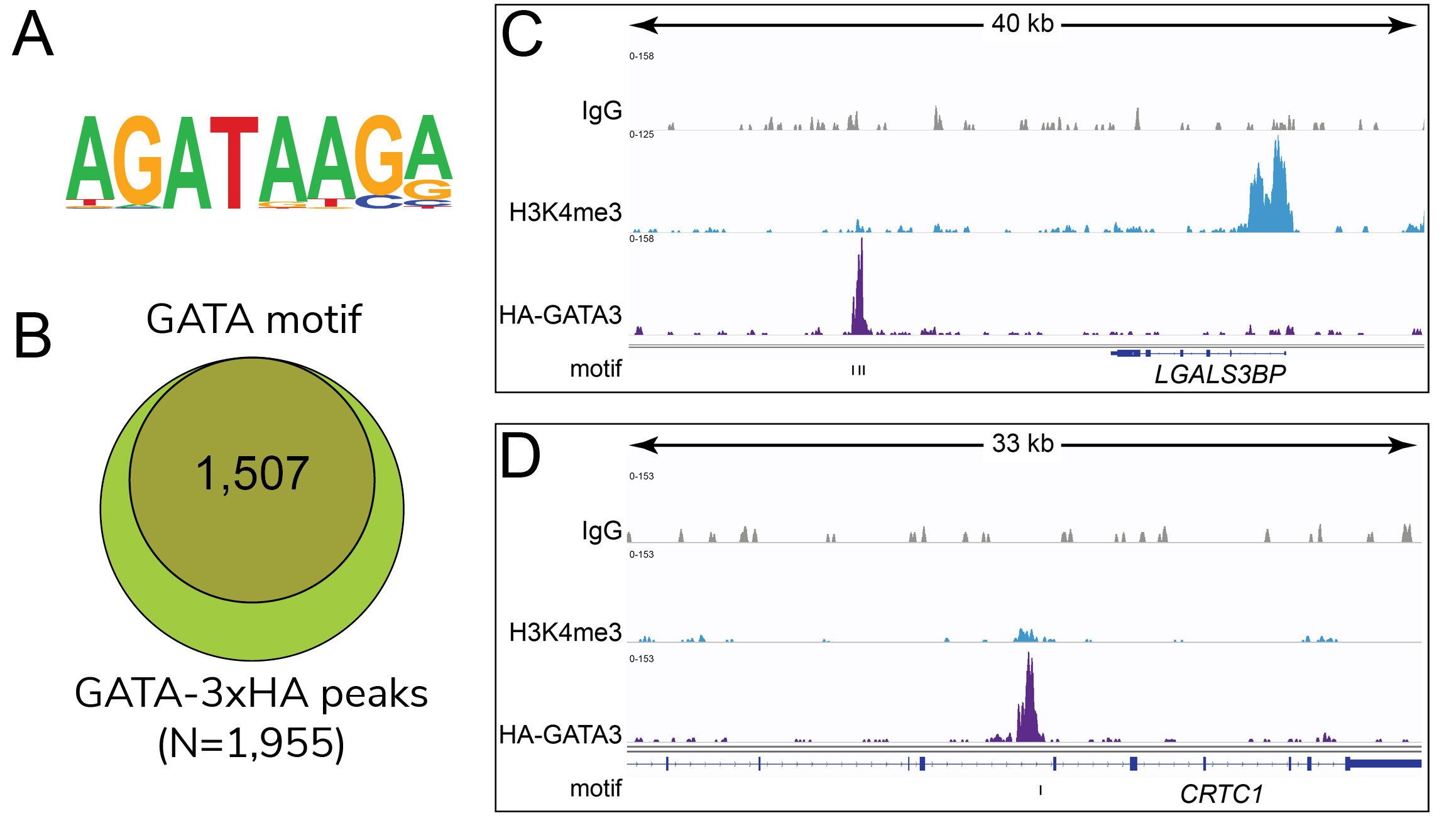
Figure 2: HA-tagged transcription factor binding
motif analysis in CUT&RUN
(A) Homer analysis determined that
the GATA3 consensus motif, represented as a sequence
logo position weight matrix, was enriched under
GATA3-3xHA CUT&RUN peaks. (B) The
number of GATA3-3xHA peaks containing GATA3 consensus
motifs from panel A is represented by a Venn Diagram.
(C-D) Two representative loci show
overlap of GATA3-3xHA peaks with the consensus motifs
noted by tick marks beneath the tracks.
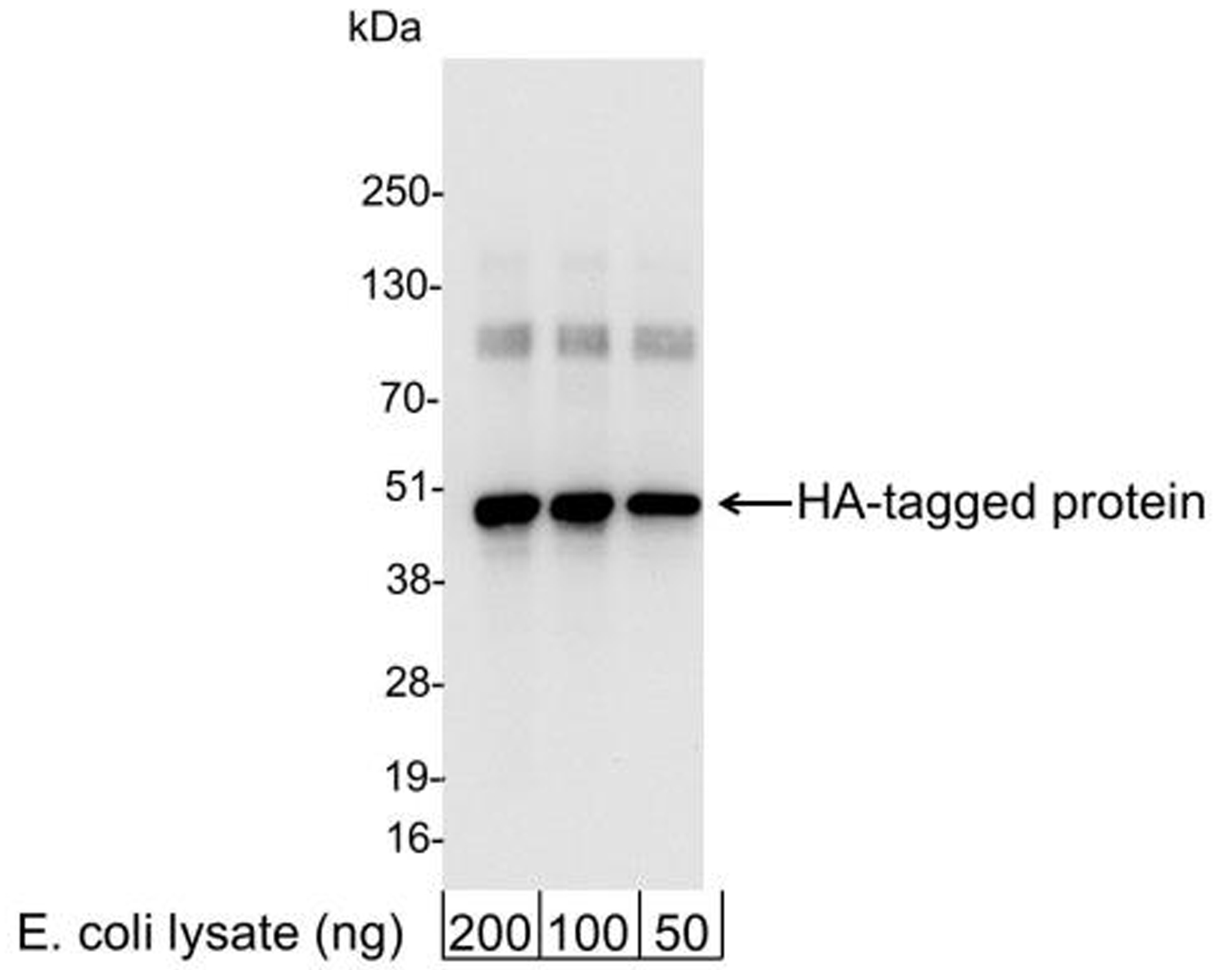
Figure 3: Western blot data
E. coli cells expressing a multi-tag fusion
protein were used to prepare whole cell lysates. The
indicated amounts (ng) of lysate were loaded onto a
4-20% SDS-PAGE gel and analyzed under standard western
blot conditions using HA Tag antibody at a dilution of
1:25,000.
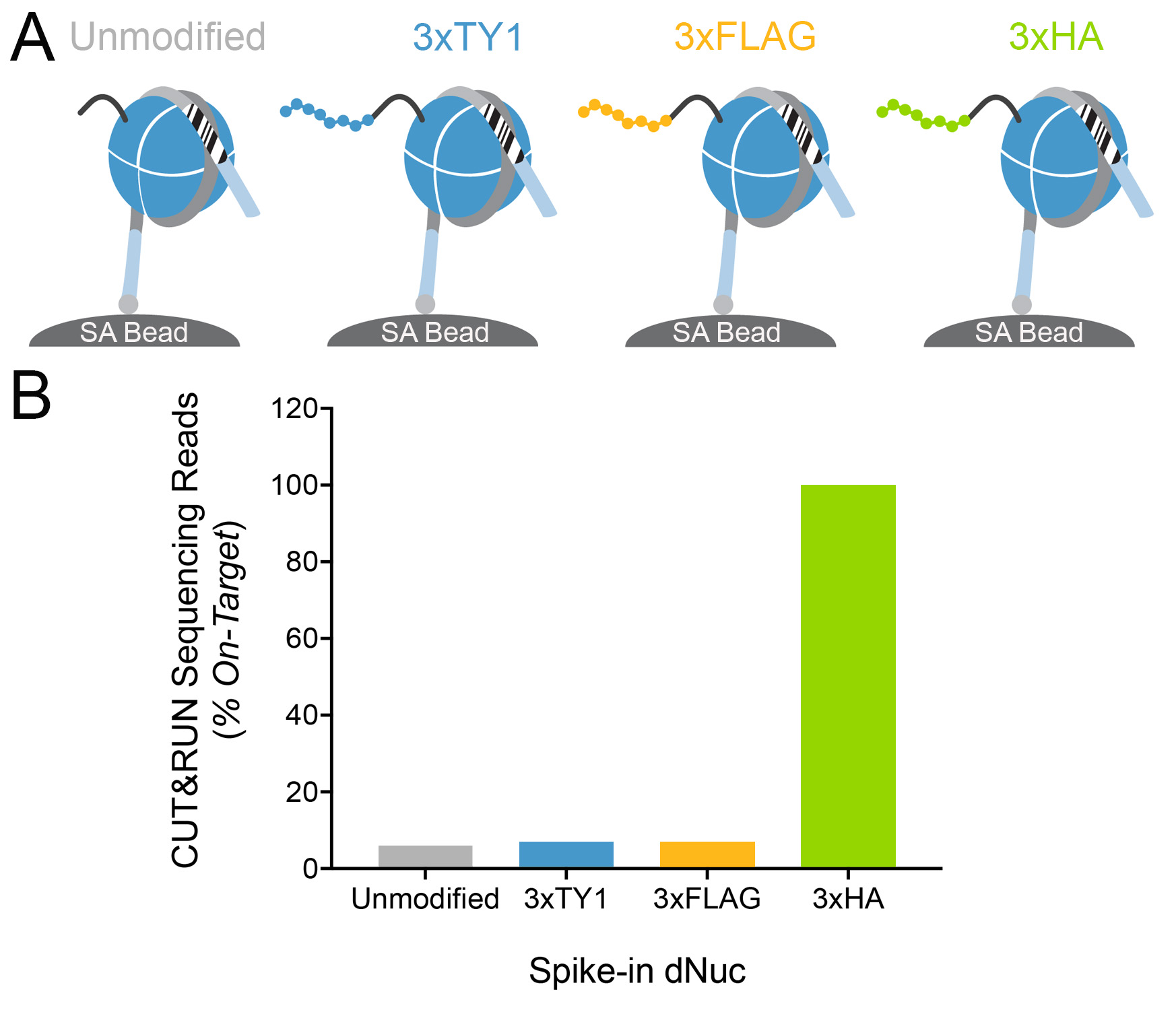
Figure 4: Target-specific epitope cleavage of HA
Tag antibody in CUT&RUN was determined using
DNA-barcoded recombinant nucleosome spike-in
controls
(A) A panel of recombinant
nucleosomes was created where various epitope tags
(3xTY1, 3xFLAG, 3xHA) were fused to the histone H3
tail. The fused nucleosomes and an unmodified control
were immobilized to streptavidin beads (SA Bead) and
spiked into CUT&RUN samples alongside ConA bead
immobilized MDA-MB-231 cells expressing GATA3-3xHA
(Figure 1). HA Tag antibody and pAG-MNase (EpiCypher
15-1016) were then added to release antibody-bound
nucleosomes into solution through pAG-MNase mediated
cleavage of the linker DNA (light blue). This approach
provided a defined experimental control to assess
whether the HA Tag antibody selectively cleaved the
target epitope with high specificity and minimal
background. (B) CUT&RUN sequence
reads were aligned to the unique DNA "barcodes"
corresponding to each nucleosome in the spike-in
panel. Data are expressed as the percent of reads
recovered relative to the intended target (3xHA, set
to 100%). This analysis confirms that the HA Tag
antibody specifically liberated the target
epitope-tagged nucleosome into solution.
Figure 5: CUT&RUN Methods
CUT&RUN was performed on 500k MDA-MB-231 cells stably
expressing c-terminal 3xHA-tagged GATA3 [1]* with 0.5
µg of either HA Tag, H3K4me3 positive control
(EpiCypher
13-0041), or IgG negative control (EpiCypher
13-0042) antibodies using the CUTANA™ ChIC/CUT&RUN Kit v2.0
(EpiCypher
14-1048). Library preparation was performed with 5 ng of DNA
(or the total amount recovered if less than 5 ng)
using the CUTANA™ CUT&RUN Library Prep Kit (EpiCypher
14-1001/14-1002). Both kit protocols were adapted for high
throughput Tecan liquid handling. Libraries were run
on an Illumina NextSeq2000 with paired-end sequencing
(2x50 bp). Sample sequencing depth was 2.4 million
reads (IgG), 4.1 million reads (H3K4me3), and 8.6
million reads (HA Tag). Data were aligned to the hg19
genome using Bowtie2. Data were filtered to remove
duplicates, multi-aligned reads, and ENCODE DAC Exclusion List
regions.
Technical Information
Recommended Dilution
Documents & Resources
Additional Info
This product is provided for commercial sale under license from Bethyl Laboratories, Inc.