CUTANA™ meCUT&RUN Kit for DNA Methylation Sequencing
In stock
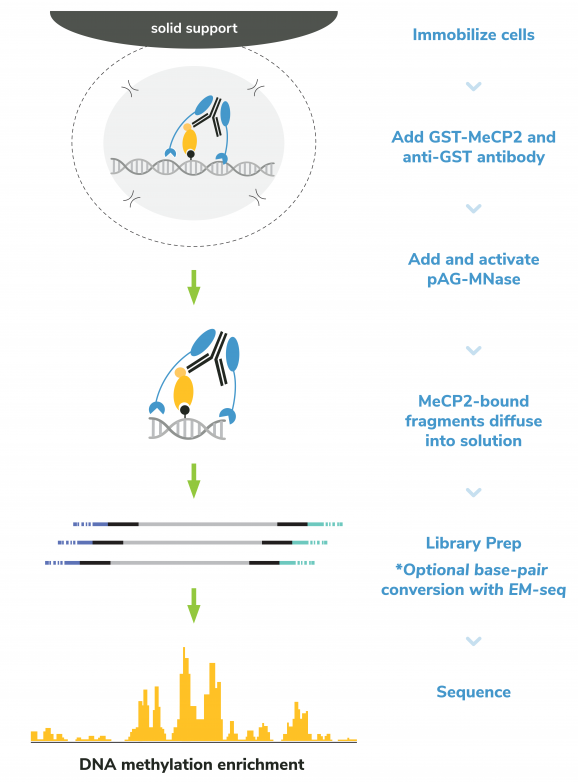
The CUTANA™ meCUT&RUN Kit for DNA Methylation Sequencing enables streamlined, high-resolution mapping of DNA methylation at >20-fold reduced sequencing depths compared to whole genome strategies like bisulfite sequencing. In meCUT&RUN, a GST-tagged MeCP2 methyl binding domain binds methylated DNA, directing the selective cleavage and release of DNA methylation-enriched chromatin fragments into solution by an immunotethered nuclease (pAG-MNase). Enriched DNA is separated from bead-bound cells, purified, and prepared for sequencing by one of two methods:
Option 1 uses a traditional library prep method, such as the CUTANA™ CUT&RUN Library Prep Kit (EpiCypher 14-1001/14-1002), to provide ~150 bp resolution profiles of DNA methylation enrichment. Only 15-20 million total sequencing reads are required for this strategy, and data are analyzed using standard CUT&RUN bioinformatic pipelines.
Option 2 requires Enzymatic Methyl-seq (NEB® EM-seq™) or alternatively bisulfite sequencing to provide base-pair resolution of 5-methylcytosine (5mC). This workflow requires 30-50 million total sequencing reads and is analyzed using standard DNA methylation bioinformatic tools; see User Manual for detailed information.
The meCUT&RUN Kit is designed for 8-strip tubes and multi-channel pipetting, enabling a seamless workflow that maximizes throughput and reproducibility. The kit is compatible with a variety of inputs including cells or nuclei derived from native, cryopreserved, or cross-linked samples. While it is recommended to start with 500,000 cells, comparable data can be generated using as few as 10,000 cells. The low sequencing depth requirements, as well as compatibility with diverse sample inputs and low cell numbers, make this kit an ideal solution for DNA methylation sequencing.
Intellectual Property
US Pat. No. 7790379, 11885814 and related patents and pending applications
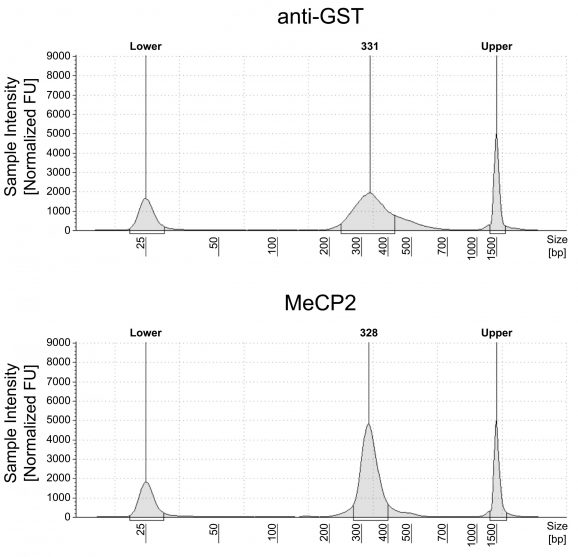
Figure 1: meCUT&RUN DNA fragment size distribution analysis
meCUT&RUN was performed as described in Figure 3. Library DNA was analyzed by Agilent TapeStation®. This analysis confirmed that mononucleosomes were predominantly enriched in meCUT&RUN (~300 bp peaks represent 150 bp nucleosomes + sequencing adapters).
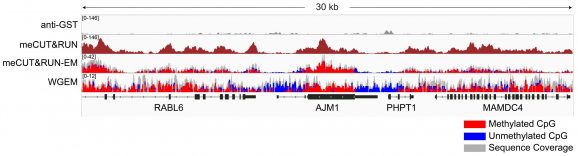
Figure 2: Gene browser tracks
meCUT&RUN was performed as described in Figure 3. A 30 kb window at the AJM1 gene is shown for anti-GST antibody and meCUT&RUN. Tracks are also shown with representative data for meCUT&RUN followed by EM-seq (meCUT&RUN-EM) and whole genome EM-seq (WGEM), using the New England Biolabs® NEBNext® Enzymatic Methyl-seq v2 Kit (NEB E8015). The meCUT&RUN kit produced the expected genomic distribution, showing enrichment of methylated DNA that approximates the methylated CpG pattern observed in WGEM. Images were generated using the Integrative Genomics Viewer (IGV, Broad Institute).
Figure 3: meCUT&RUN methods
meCUT&RUN was performed using the CUTANA™ meCUT&RUN Kit for DNA Methylation Sequencing starting with 500k K562 cells with either 2.5 µL of GST-MeCP2 (EpiCypher 15-2002) added as the primary binding reagent or 0.5 µg of a secondary antibody-only control (anti-GST antibody, EpiCypher 13-0073) added to determine background cleavage. Library preparation was performed using 5 ng of meCUT&RUN-enriched DNA (or the total amount recovered if less than 5 ng) using the CUTANA™ CUT&RUN Library Prep Kit (EpiCypher 14-1001/14-1002). Libraries were run on an Illumina NextSeq2000 with paired-end sequencing (2×50 bp). Sample sequencing depth was 7.7 million reads (anti-GST) and 8.4 million reads (GST-MeCP2). Data were aligned to the hg38 genome using Bowtie2. Data were filtered to remove duplicates, multi-aligned reads, and ENCODE DAC Exclusion List regions.
Pack Size
24 Reactions
Storage
OPEN KIT IMMEDIATELY and store components at room temperature, 4°C, and -20°C as indicated (see User Manual corresponding to Kit Version 1). Stable for 12 months upon date of receipt.
Instructions for Use
See User Manual corresponding to Kit Version 1.
| Item | Cat. No. |
|---|---|
| CUTANA™ GST-MeCP2 for meCUT&RUN | 15-2002-04 |
| CUTANA™ Anti-GST Tag Antibody | 13-0073-04 |
| CUTANA™ pAG-MNase for ChIC/CUT&RUN Workflows | 15-1016-01 |
| CUTANA™ Concanavalin A Conjugated Paramagnetic Beads | 21-1401-01 |
| CUTANA™ E. coli Spike-in DNA | 18-1401-01 |
| CUTANA™ Bead Activation Buffer | 21-1001-01 |
| CUTANA™ Stop Buffer | 21-1003-01 |
| CUTANA™ 5% Digitonin | 21-1004-01 |
| CUTANA™ CUT&RUN 8-Strip 0.2 mL Tubes | 10-0009-01 |
| Pre-Wash Buffer | 21-1002-01 |
| 1 M Spermidine | 21-1005-01 |
| 0.5 M EDTA | 21-1006-01 |
| 100 mM Calcium Chloride | 21-1007-01 |
| 5 M NaCl | 21-1013-04 |
| 0.1X TE Buffer | 21-1025-01 |
| SPRIselect reagent manufactured by Beckman Coulter, Inc. | 21-1405-01 |
| Item | Cat. No. |
|---|---|
| Option 1: CUTANA™ CUT&RUN Library Prep Kit | 14-1001/14-1002 |
| Option 2: New England Biolabs® NEBNext® Enzymatic Methyl-seq v2 Kit | NEB E8015 |
| Magnetic Separation Rack, 0.2 mL Tubes | 10-0008 |
| Magnetic Separation Rack, 1.5 mL Tubes | 10-0012 |
| CUTANA™ Nuclei Extraction Buffer | 21-1026 |
| CUTANA™ Protease Inhibitor Tablets | 21-1027 |
| CUTANA™ Quick Cleanup DNA Purification Kit | 14-0052 |