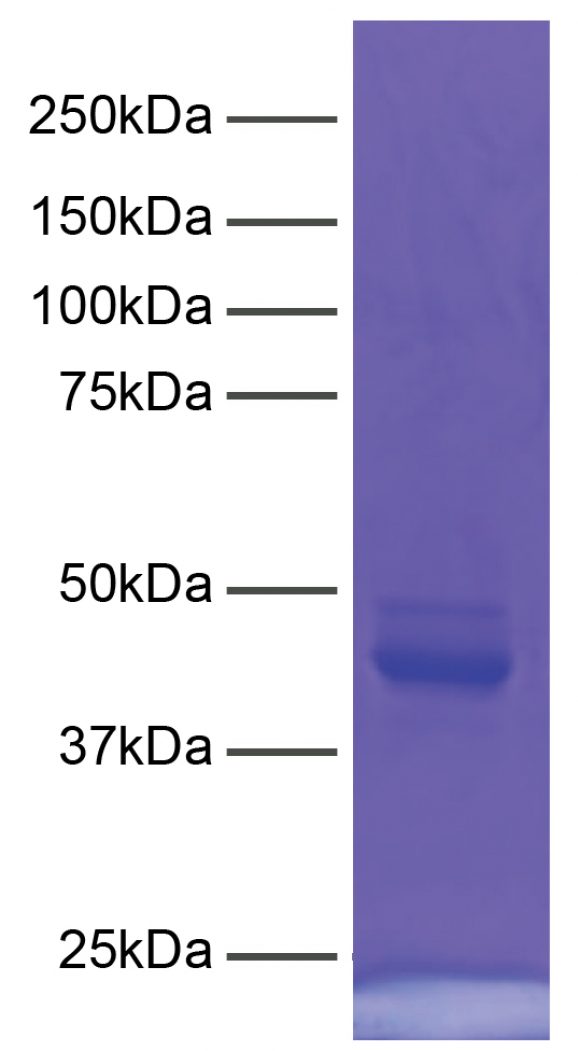
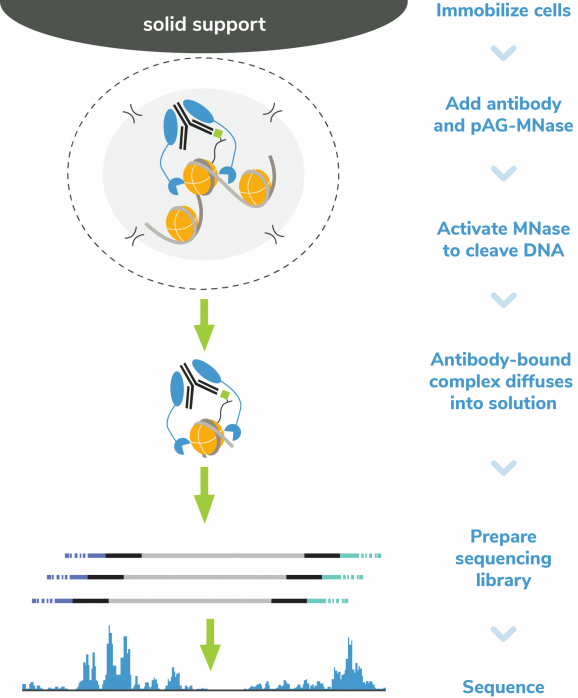
CUTANA™ pAG-MNase for ChIC/CUT&RUN Workflows
CUTANA™ pAG-MNase is the key reagent for performing Chromatin Immunocleavage (ChIC) [Schmid et al., 2004] and Cleavage Under Targets and Release Using Nuclease (CUT&RUN) [Skene & Henikoff, 2017, Skene et al., 2018].
As a fusion of Proteins A and G to Micrococcal Nuclease, CUTANA pAG-MNase is compatible with target antibodies from a broad spectrum of host species and is highly purified to remove contaminating E. coli DNA, which can complicate analysis at low cell numbers. This enzyme enables efficient mapping of chromatin features in ChIC/CUT&RUN, allowing for significant improvements in signal to noise at reduced cell inputs and sequencing depth compared to ChIP-seq.
Intellectual Property
US Pat. No. 7790379, 11885814 and related patents and pending applications
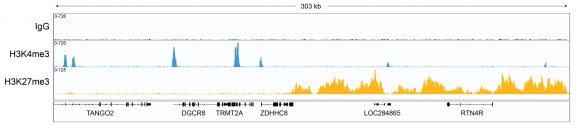
Figure 1: CUT&RUN gene browser tracks
CUT&RUN was performed as described above. Data verifies low non-specific MNase digestion with the absence of peaks in the IgG track, an expected H3K4me3 profile with sharp promoter peaks, and broad peaks in heterochromatin regions consistent with H3K27me3. Image was generated using the Integrative Genomics Viewer (IGV, Broad Institute).
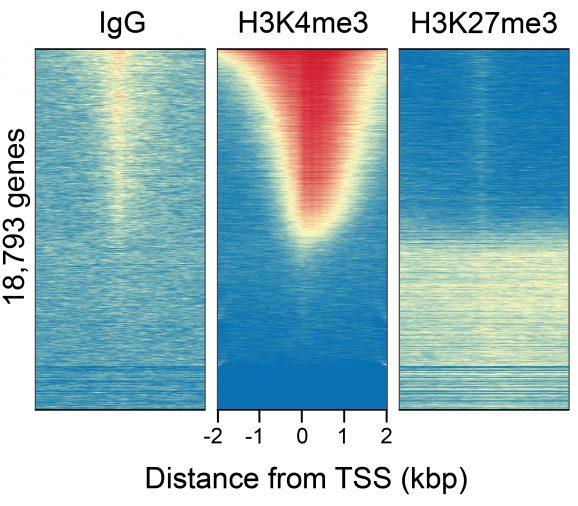
Figure 3: CUT&RUN genome-wide heatmaps
CUT&RUN was performed as described above. Heatmaps show CUT&RUN signal aligned to annotated transcription start sites (TSS, +/- 2kb). High and low signal are ranked by intensity (top to bottom) and colored such that red indicates high localized enrichment and blue denotes background signal. Gene rows in each heatmap are aligned and sorted from high to low signal relative to H3K4me3 (middle).
CUT&RUN methods
CUT&RUN was performed on 500k K562 cells with 0.5 µg of either IgG (EpiCypher 13-0042), H3K4me3 (EpiCypher 13-0041), or H3K27me3 (ThermoFisher MA5-11198) antibodies using CUTANA™ pAG-MNase (1:20 dilution) and the CUTANA™ ChIC/CUT&RUN Kit v3 (EpiCypher 14-1048). Library preparation was performed with 5 ng of DNA (or the total amount recovered if less than 5 ng) using the CUTANA™ CUT&RUN Library Prep Kit (EpiCypher 14-1001/14-1002). Both kit protocols were adapted for high throughput Tecan liquid handling. Libraries were run on an Illumina NextSeq2000 with paired-end sequencing (2×50 bp). Sample sequencing depth was 3.6 million reads (IgG), 4.3 million reads (H3K4me3), and 5.2 million reads (H3K27me3). Data were aligned to the hg19 genome using Bowtie2. Data were filtered to remove duplicates, multi-aligned reads, and ENCODE DAC Exclusion List regions.
Storage
Stable for one year at -20°C from date of receipt. The protein is not subject to freeze/thaw under these conditions.
Add 2.5 µL of the supplied enzyme to a 50 µL CUT&RUN reaction (20X dilution). For detailed applications and uses of this product, please see our CUT&RUN protocol.
| Item | Cat. No. |
|---|---|
| CUTANA™ CUT&RUN Library Prep Kit | 14-1001 |
| CUTANA™ DNA Purification Kit | 14-0050 |
| CUTANA™ Concanavalin A Conjugated Paramagnetic Beads | 21-1401 |
| SNAP-CUTANA™ K-MetStat Panel | 19-1002 |
| CUT&RUN Antibodies | See the list |
| CUTANA™ E. coli Spike-in DNA | 18-1401 |
| Magnetic Separation Rack, 0.2 mL Tubes | 10-0008 |
| Magnetic Separation Rack, 1.5 mL Tubes | 10-0012 |
| CUTANA™ CUT&RUN 8-strip 0.2 mL Tubes | 10-0009 |
| CUTANA™ Nuclei Extraction Buffer | 21-1026 |
| CUTANA™ Protease Inhibitor Tablets | 21-1027 |
Need help with bioinformatic analysis?
Go from raw data to insights in just a few clicks with CUTANA® Cloud.
Get Started ›