Multiomics Made Easy with CUTANA™ Multiomic CUT&RUN
- Aaron Alcala, PhD

Gene expression is regulated by the complex molecular interplay between DNA methylation and chromatin proteins (e.g., transcription factors, post-translational modifications [PTMs]). Traditionally, researchers have studied these genomic features separately using methods like ChIP-seq and bisulfite sequencing — approaches that are expensive, time-consuming, and require complex data integration.
CUTANA™ Multiomic CUT&RUN enables true multiomic profiling by simultaneously mapping chromatin proteins and DNA methylation in a single assay. This powerful approach offers a new lens for studying complex chromatin signaling by revealing how different gene regulatory layers intersect. Multiomic CUT&RUN makes it easier than ever to explore the crosstalk between DNA methylation and chromatin proteins such as histone PTMs, transcription factors, and chromatin-modifying enzymes.
Why is Multiomic CUT&RUN better for simultaneous profiling of DNA methylation and chromatin proteins?
Traditional multiomic DNA methylation approaches combine chromatin immunoprecipitation sequencing (ChIP-seq) with whole-genome bisulfite sequencing (WGBS) datasets (Figure 1). However, these workflows are cost-prohibitive, require large numbers of cells, and take weeks (or longer!) to complete. ChIP-seq also uses a complicated workflow that is expensive and produces high background. Moreover, simply overlaying ChIP-seq and WGBS datasets cannot directly capture how chromatin proteins and DNA methylation functionally interact at the same loci, limiting the biological insights that can be achieved.
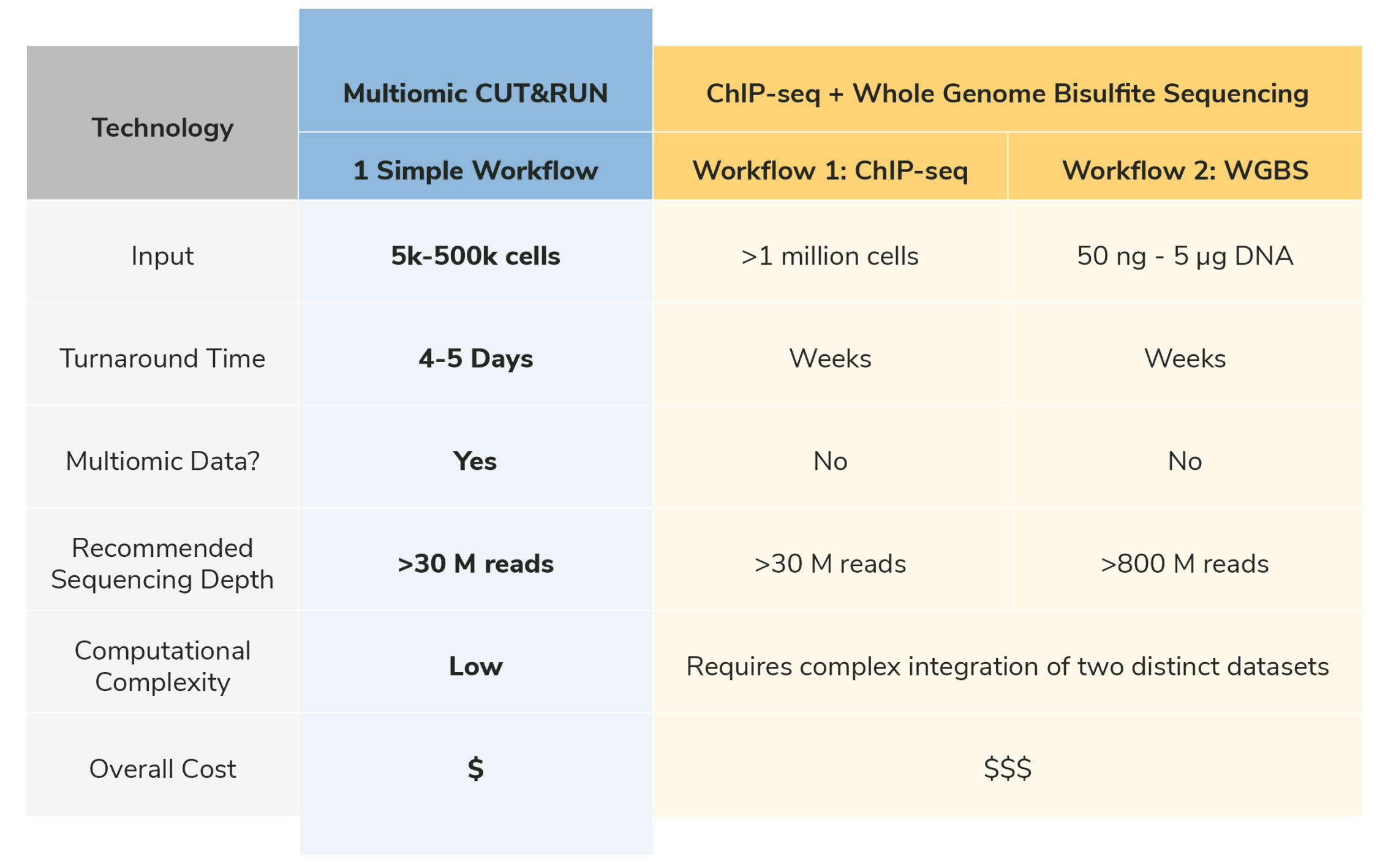
Figure 1: Multiomic CUT&RUN provides deep insight into chromatin regulation in a single workflow.
Multiomic CUT&RUN streamlines this process by generating DNA methylation profiles specifically at protein-bound regions, with a fraction of the time, input, and cost required for ChIP-seq and WGBS. This powerful approach produces high-quality genomic maps for transcription factor binding sites, histone post-translational modifications (PTMs), and chromatin-modifying enzymes, while simultaneously revealing differential DNA methylation patterns, such as hypermethylation at gene bodies and hypomethylation at promoters (Figure 2).
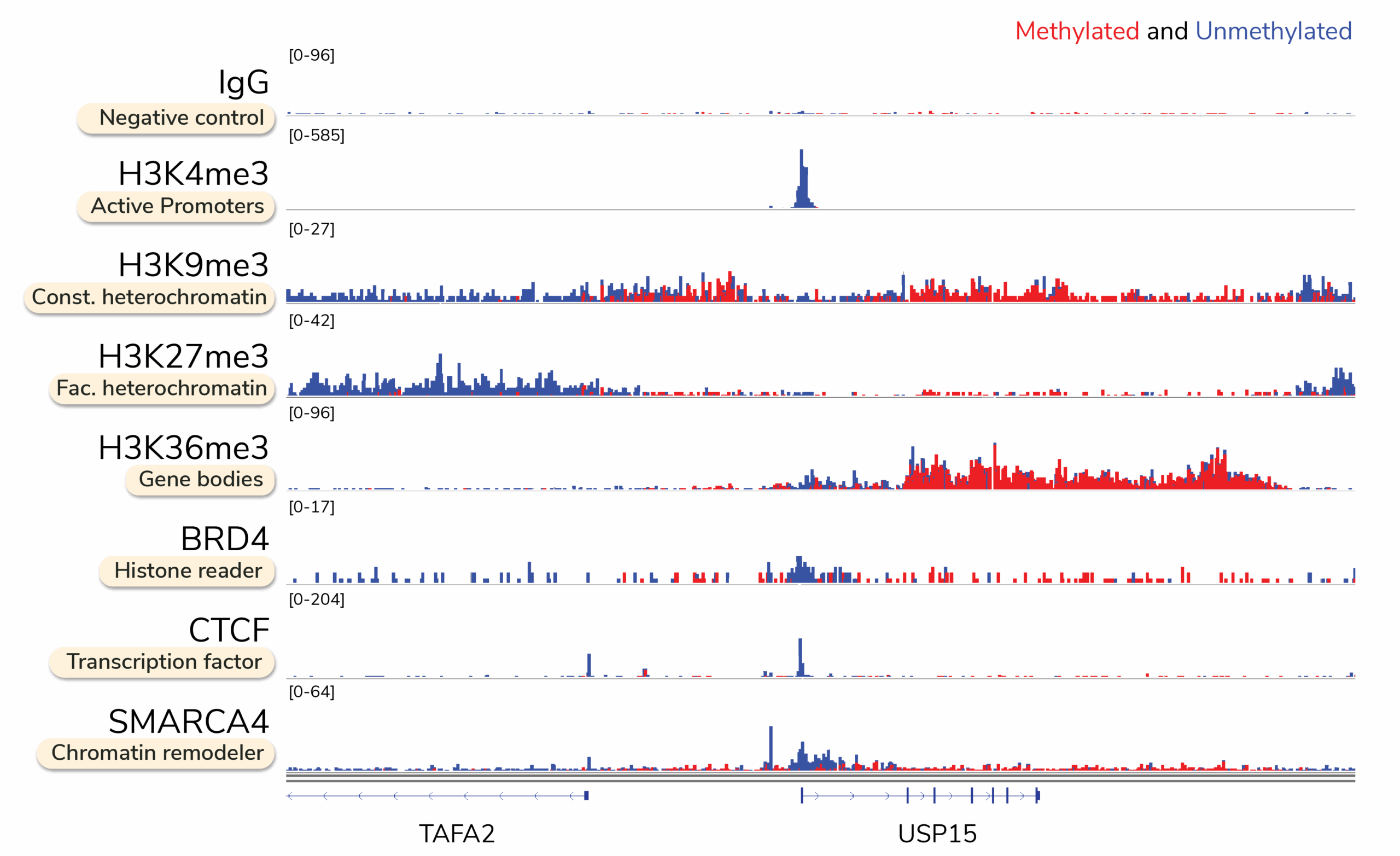
Figure 2: Multiomic CUT&RUN provides base-pair resolution of CpG methylation at transcription factor binding sites, histone PTMs, and chromatin-modifying enzymes. Each reaction used 500,000 K562 cells. Target chromatin was enriched with the Multiomic CUT&RUN Workflow, followed by Enzymatic Methyl-seq (EM-seq) conversion and library prep.
How does Multiomic CUT&RUN work?
CUTANA™ Multiomic CUT&RUN maps DNA methylation at protein binding sites. This assay leverages CUT&RUN technology to first enrich for antibody-bound chromatin (Figure 3).
Next, Enzymatic Methyl-seq (NEB® EM-seq™, preferred*) or bisulfite sequencing is performed to convert unmethylated cytosines to uracils for base-pair resolution analysis of DNA methylation. This streamlined approach requires only 30-50 million total reads for robust multiomic analysis of protein binding sites and methylated CpG sites (5mCs). Alternatively, bisulfite conversion can be performed to achieve base-resolution methylation mapping.
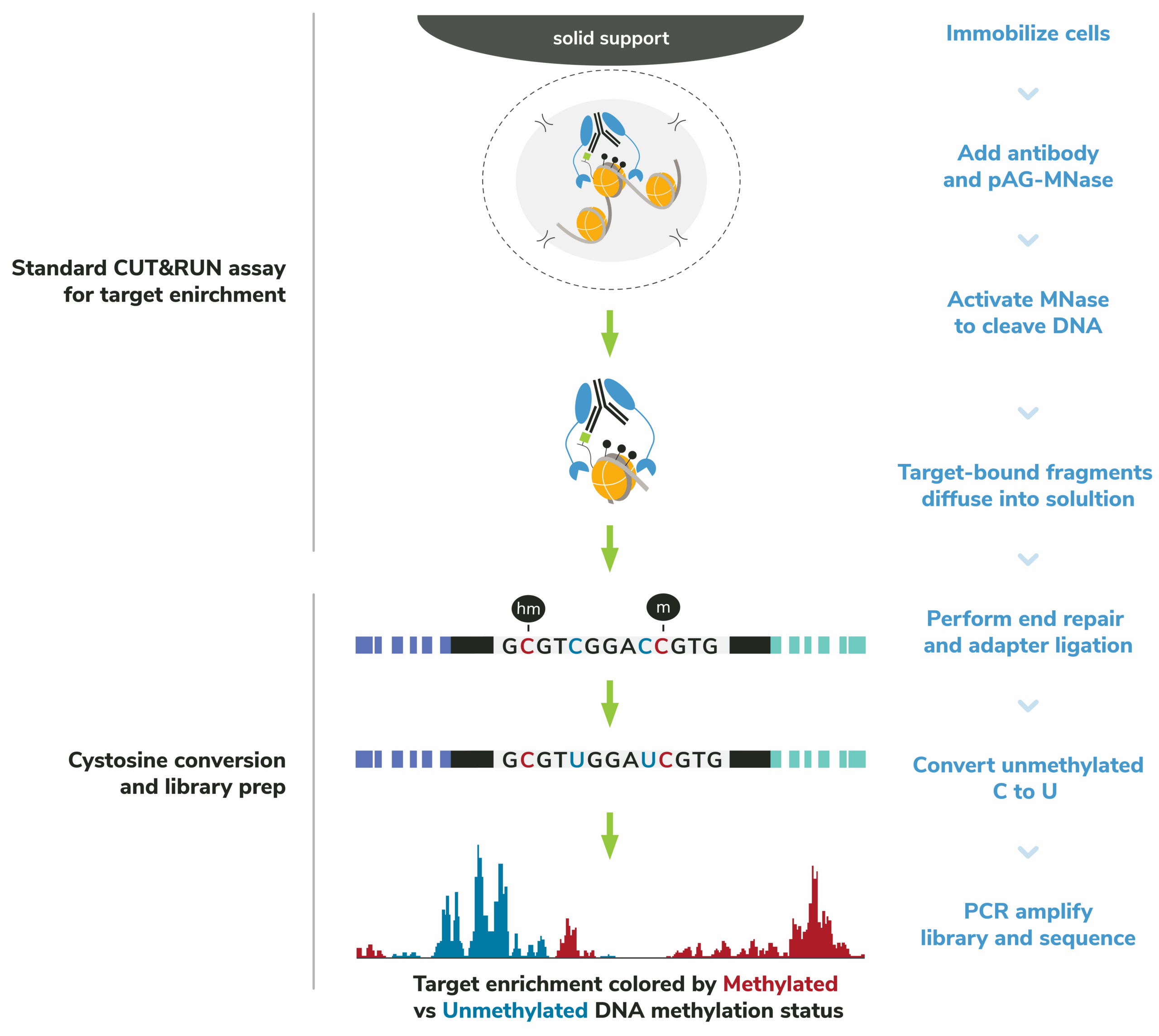
Figure 3: The CUTANA™ Multiomic CUT&RUN workflow. First, CUT&RUN is used to enrich target-bound chromatin. Only 5,000-500,000 cells per reaction is needed. Next, unmethylated cytosines are converted to uracils for base-pair resolution DNA methylation analysis. Note that enzymatic or bisulfite conversion may be used. Lastly, next-generation sequencing and data analysis is performed. Only 30-50 M total reads are needed for robust multiomic analysis of target protein and underlying DNA methylation.
Get Started with CUTANA™ Multiomic CUT&RUN Today!
Ready to bring your DNA methylation research to the next level? We offer a collection of Multiomic CUT&RUN reagents:
- Multiomic CUT&RUN Controls Set: A specialized set of controls for Multiomic CUT&RUN, as well as optimized spike-ins for DNA methylation conversion.
- CUTANA™ CUT&RUN Kit: A user-friendly kit to enrich target chromatin; key for Multiomic CUT&RUN assays.
- CUT&RUN-Validated Antibodies: High quality antibodies with exquisite sensitivity, expertly validated for CUT&RUN.
* NEB®, New England Biolabs® and NEBNext® are registered trademarks of New England Biolabs, Inc. EM-Seq™ is a trademark of New England Biolabs, Inc.
