CUTANA™ Fiber-seq Kit
The CUTANA™ Fiber-seq Kit provides key reagents and a comprehensive manual to perform Fiber-seq. Key advantages include:
- Multiomic insights in one assay – Fiber-seq simultaneously profiles chromatin accessibility, DNA methylation, and genetic variants, consolidating what typically requires three or more separate assays into one long-read sequencing (LRS) assay [1].
- Comprehensive solution – This kit includes key reagents needed to go from cells to library prep-ready DNA.
- User-friendly resources – The User Manual is complete with a detailed protocol, QC checks, FAQs, and expert tips to get you started, while the bench-proof Quick-Start Card provides a concise protocol for convenient reference.
- Easy workflow integration – Fiber-seq can be performed in <2 hours directly upstream of routine long-read sequencing library preparation and uses reagents vetted for compatibility with library prep kits from major LRS platforms.
- Powered by Hia5 6mA methyltransferase – Hia5 is the highest activity 6mA-methyltransferase available and is exclusive to CUTANA Fiber-seq Kits. This more active enzyme enables faster incubations, which results in better resolution of protein footprints [2-4].
Stringent quality control – Each new lot undergoes rigorous quality control checks to ensure components yield consistent results in your research.
Fiber-seq is a breakthrough approach that simultaneously maps genetic and epigenetic features from individual chromatin fibers. This single assay delivers insights that would normally require integrating data from multiple methods such as ATAC-seq, ChIP-seq, and bisulfite sequencing. This kit includes reagents to go from cells to purified genomic DNA that can be used directly in either Pacific Biosciences® (PacBio® HiFi Sequencing) or Oxford Nanopore Technologies® (ONT® Nanopore Sequencing) library preparation workflows. The recommended input for Fiber-seq is 1,000,000 native human nuclei per reaction. The Fiber-seq Kit has been successfully applied to cryopreserved and lightly cross-linked nuclei (refer to the Tech Support Center for the most up-to-date recommendations).
Intellectual Property
Fiber-seq is a patent-pending technology.
Relevant pending applications include PCT/US2020/017597, PCT/US2021/025644, and related patents and patent applications.
Pacific Biosciences, PacBio, Revio, and SMRT are registered trademarks of Pacific Biosciences of California, Inc.
Oxford Nanopore Technologies and ONT are registered trademarks of Oxford Nanopore Technologies Limited.
All other trademarks are the property of their respective owners.
Figure 1: The Fiber-seq workflow
In the Fiber-seq workflow, isolated nuclei are incubated with Hia5 N6-methyladenine methyltransferase (6mA MTase) and cofactor S-Adenosylmethionine (SAM) to selectively methylate adenines within regions of accessible chromatin. After a brief 10-minute labeling step, the reaction is quenched and genomic DNA (gDNA) is purified and prepared for long-read sequencing following standard native DNA sequencing protocols. Long-read sequencing data is next analyzed at a per molecule level to observe heterogeneity in chromatin structure within a sample or in aggregate to identify sample-wide trends in accessibility and DNA methylation.
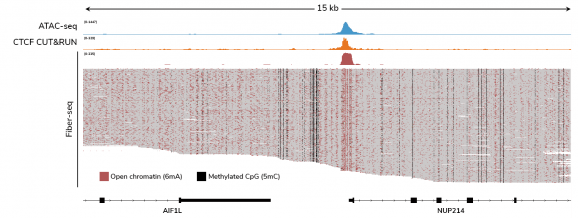
Figure 2: Fiber-seq simultaneously captures chromatin accessibility and endogenous DNA methylation
CUTANA™ Fiber-seq simultaneously detects open chromatin (marked by N6-methyladenine; 6mA) and DNA methylation (5mC) in human leukemia (K562) cells. The top Fiber-seq track displays aggregate 6mA signal. ATAC-seq and CTCF CUT&RUN data are shown for comparison. CTCF is a DNA-binding protein that regulates chromatin structure and is often enriched at open chromatin regions. Each horizontal line in the Fiber-seq panel represents a single long read, providing single-DNA molecule data from 60 individual chromatin fibers.
Figure 3: Fiber-seq reveals protein footprints genome wide and at higher resolution than ATAC-seq
Figure 4: Fiber-seq captures chromatin accessibility, DNA methylation, and inferred nucleosome positioning genome-wide from a single experiment
Heatmaps show aligned single-molecule data for 6mA signal, nucleosome positioning, and endogenous 5mC levels centered at transcription start sites (TSSs). TSSs enriched for 6mA display well-defined nucleosome organization and a depletion of 5mC, consistent with active promoter regions. These patterns align with CUT&RUN profiles for histone modifications, including H3K4me3 (active TSS) and H3K27me3 (repressed genes).
Storage
OPEN KIT IMMEDIATELY and store components at room temperature, 4°C, -20°C, and -80°C as indicated (see Kit Version 1 User Manual). Stable for 6 months upon date of receipt.
Instructions for Use
See User Manual corresponding to Kit Version 1.
| Item | Cat. No. |
|---|---|
| CUTANA™ Hia5 For Fiber-seq | 15-1032 |
| CUTANA™ Fiber-seq Kit SAM | 21-1034 |
| CUTANA™ Fiber-seq Kit Pre-1X Reaction Buffer | 21-1033 |
| CUTANA™ Pre-Nuclei Extraction Buffer | 21-1021 |
| 1 M Spermidine | 21-1005 |
| Proteinase K | 15-1033 |
| RNase A | 15-1034 |
| gDNA Spin Columns | 10-0016 |
| gDNA Collection Tubes | 10-0017 |
| gDNA Cell Lysis Buffer | 21-1029 |
| gDNA Binding Buffer | 21-1030 |
| gDNA Wash Buffer | 21-1031 |
| gDNA Elution Buffer | 21-1032 |