BRM/SMARCA2 CUTANA™ CUT&RUN Antibody
In stock
This antibody meets EpiCypher’s “CUTANA Compatible” criteria for performance in Cleavage Under Targets and Release Using Nuclease (CUT&RUN) and/or Cleavage Under Targets and Tagmentation (CUT&Tag) approaches to genomic mapping. Every lot of a CUTANA Compatible antibody is tested in the indicated approach using EpiCypher optimized protocols and determined to yield peaks that show a genomic distribution pattern consistent with reported function(s) of the target protein. BRM/SMARCA2 antibody produces CUT&RUN peaks above background (Figure 1) localized to gene transcription start sites (Figures 1-2), consistent with its known role as the ATP-dependent helicase subunit of the SWI/SNF chromatin remodeler complex [1].
Trademarks and copyrights
This product is provided for commercial sale under license from Bethyl Laboratories, Inc.
Background References:
[1] Raab et al. Epigenetics Chromatin (2017). PMID: 29273066
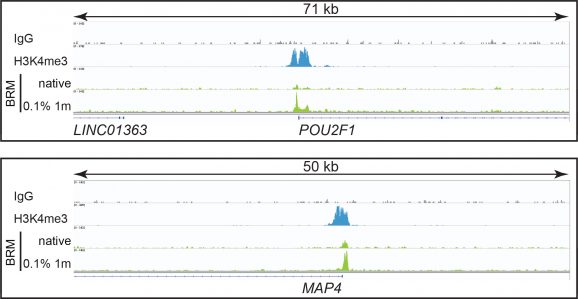
CUT&RUN was performed on 500k native or fixed (0.1% formaldehyde, 1 min) K562 cells with 0.5 µg of either BRM, H3K4me3 positive control (EpiCypher 13-0041), or IgG negative control (EpiCypher 13-0042) antibodies using the CUTANA™ ChIC/CUT&RUN Kit v2.0 (EpiCypher 14-1048). Library preparation was performed with 5 ng of DNA (or the total amount recovered if less than 5 ng) using the CUTANA™ CUT&RUN Library Prep Kit (EpiCypher 14-1001/14-1002). Both kit protocols were adapted for high throughput Tecan liquid handling. Libraries were run on an Illumina NextSeq2000 with paired-end sequencing (2×50 bp). Sample sequencing depth was 10.2 million reads (BRM native), 10.0 million reads (BRM 0.1% fixation), 4.1 million reads (H3K4me3), and 2.4 million reads (IgG). Data were aligned to the hg19 genome using Bowtie2. Data were filtered to remove duplicates, multi-aligned reads, and ENCODE DAC Exclusion List regions.
Figure 1: BRM peaks in CUT&RUN
Figure 2: BRM CUT&RUN representative browser tracks
Figure 4: Immunoprecipitation data
- Type: Polyclonal
- Host: Rabbit
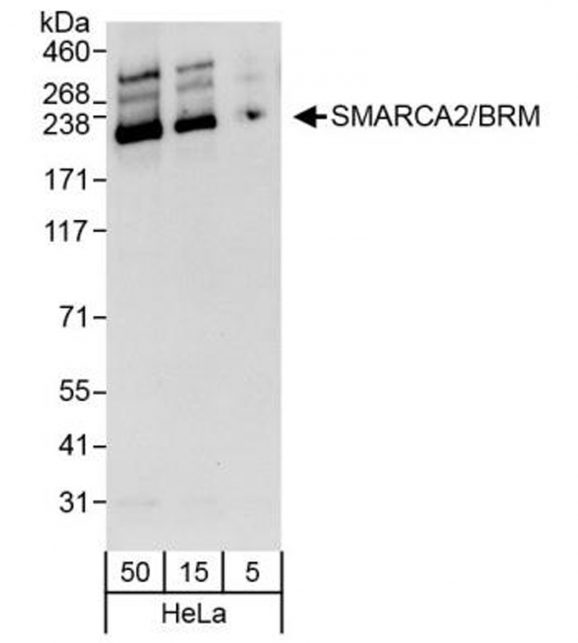
- Applications: CUT&RUN, WB, IP
- Reactivity: Human
- Format: Antigen affinity-purified
- Target Size: 181 kDa
- CUT&RUN: 0.5 µg per reaction
- Western Blot (WB): 1:2,000 – 1:10,000
- Immunoprecipitation (IP): 2 – 5 µg/mg lysate
- UniProt ID: P51531
- Gene Name: SMARCA2
- Protein Name: Probable global transcription activator SNF2L2
- Target Size: 181 kDa
- Alternate Names: BAF190B, SNF2A, SNF2L2, ATP-dependent helicase SMARCA2, BRG1-associated factor 190B (BAF190B), Protein bahma homolog (hBRM), SNF2-alpha, SWI-SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 2