CUTANA™ DNA Methylation
Sequencing Services
Your partner for premium methylation mapping services
DNA methylation is a stable epigenetic mark that plays a central role in gene regulation, cell identity, and disease progression. As a readout of regulatory state, it provides insights into drug target engagement, therapeutic response, and drug resistance mechanisms.
By capturing how regulatory programs are established, maintained, or disrupted, DNA methylation serves as a powerful marker for understanding drug mechanism of action and discovering biomarkers. Importantly, these regulatory programs — and the phenotypes they produce — emerge from the interplay between DNA methylation, chromatin accessibility, transcription factor binding, histone modifications, and higher-order genome organization. Understanding how these layers change together is essential for translating discoveries into clinical impact.
EpiCypher provides next-generation tools for DNA methylation sequencing, offering innovative alternatives to destructive bisulfite-based methods. Our services provide unique access to our genomic expertise, enabling diverse applications across biomedical research and drug development. As our partner, you can expect:
- Detailed, end-to-end experimental design
- Options for whole-genome or targeted DNA methylation profiling, as well as long-read multiomic studies
- Expert optimization for targets, cell types, tissues, and clinical samples
- A comprehensive data report and presentation by our scientists
- Scalable protocols from pilot tests to high-throughput studies

Have Questions?
We’re here to help. Click below and a member of our team will get back to you shortly!
Find the Right DNA Methylation Profiling Assay for Your Project

Genome-Wide DNA Methylation Mapping
Powered by meCUT&RUN, this approach enables scalable whole-genome DNA methylation profiling while requiring ~20-fold less sequencing than whole-genome bisulfite sequencing. This makes meCUT&RUN ideally suited for discovery and perturbation studies.

Targeted DNA Methylation Profiling
Multiomic CUT&RUN profiles DNA methylation directly at targeted chromatin proteins, enabling causal linkage between methylation changes and regulatory element activity. This integrated approach is well suited for mechanism-of-action studies, where understanding how DNA methylation influences chromatin regulation is essential.
CUTANA™ Fiber-seq: long-read multiomics
Fiber-seq provides a comprehensive view of gene regulation by simultaneously profiling chromatin accessibility, DNA methylation, protein footprints, and genetic variation in a single long-read sequencing assay. By delivering single-molecule resolution, Fiber-seq reveals epigenomic insights in complex and heterogeneous samples that bulk assays cannot resolve.
Not sure which assay is right for you?
Start with our blog on DNA methylation mapping methods, or contact our team for personalized guidance.
Tell us about your project:
FAQs about our DNA Methylation Sequencing Services
What are the differences between the DNA methylation assay options?
Each assay provides a different level of insight depending on your project needs:
- meCUT&RUN is a cost-effective CUT&RUN-based method for genome-wide 5mC profiling, delivering broad CpG coverage with low sequencing requirements. We offer two library prep formats: standard meCUT&RUN, which provides an overview of 5mC enrichment throughout the genome, and meCUT&RUN-EM, which uses enzymatic conversion to deliver base-pair–resolution methylation maps.
- Multiomic CUT&RUN goes a step further by mapping DNA methylation together with chromatin proteins (e.g., transcription factors, histone PTMs), enabling base-resolution methylation analysis specifically at targeted protein binding sites.
- Fiber-seq provides a long-read, single-molecule view of native DNA methylation, chromatin accessibility, and genetic variation in one experiment—ideal for resolving complex regions, allele-specific regulation, and chromatin structure at unprecedented resolution.
How do I decide which DNA methylation assay to choose?
Our team brings deep expertise in epigenetics and functional genomic assays. We work directly with you to design the right DNA methylation profiling strategy for your biological question, sample type, and stage of development. You bring the biology — we help identify the solution that best fits your needs.
In practice, we leverage a suite of DNA methylation assays depending on the depth of insight required:
meCUT&RUN for scalable, cost-effective whole-genome methylation profiling in discovery and perturbation studies.
Multiomic CUT&RUN to link DNA methylation directly to chromatin regulatory elements for mechanism-of-action studies.
Fiber-seq for a comprehensive view of DNA methylation, open chromatin, protein footprints, and genetic variants in a single long-read assay — ideal for resolving complex or heterogeneous samples.
Not sure which option is right for your project? Reach out to our team using the form above—we’d be happy to help you decide.
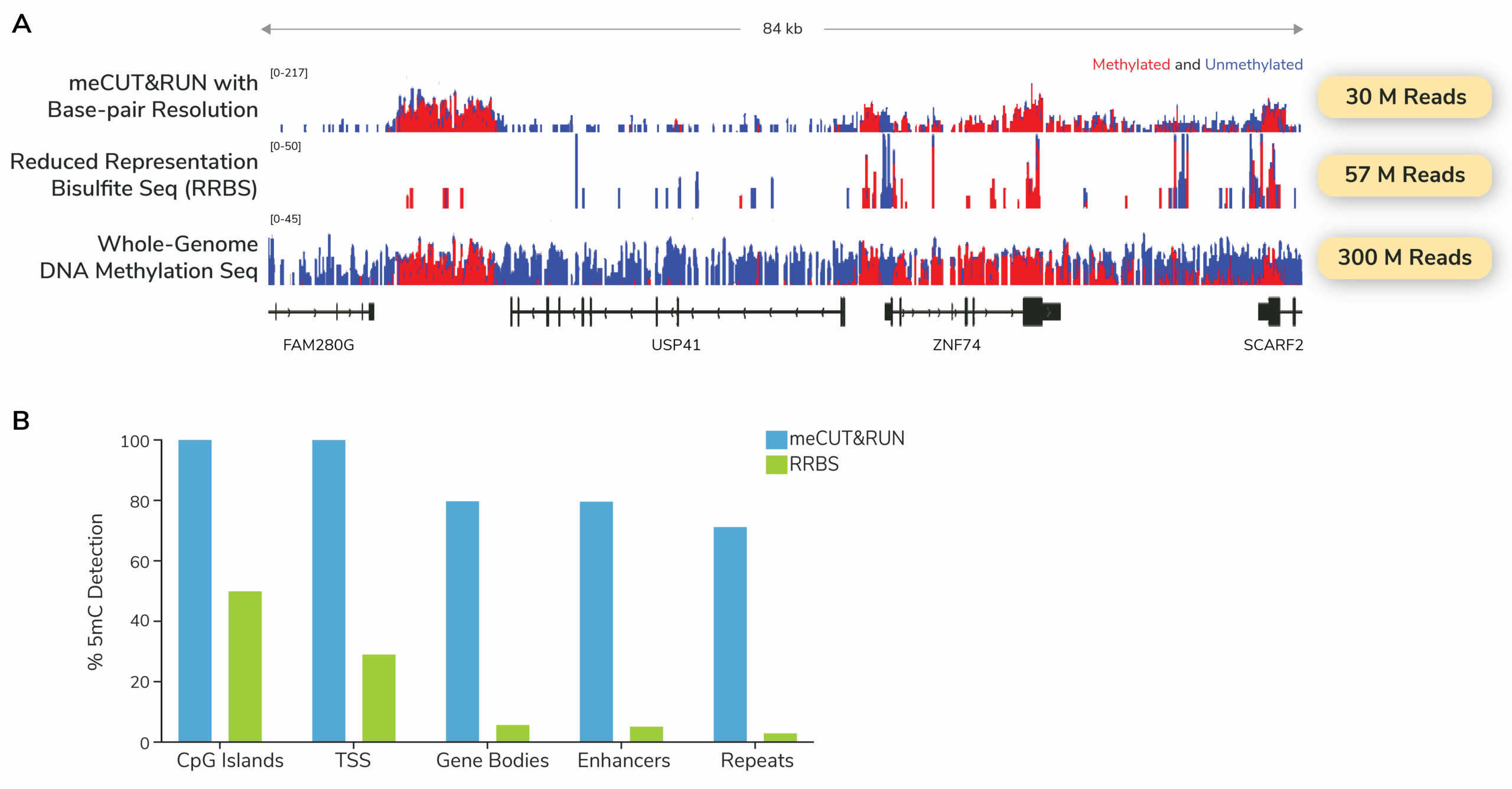
What does whole-genome methylation sequencing data look like with meCUT&RUN?
The figure below compares meCUT&RUN to Reduced Representation Bisulfite Sequencing (RRBS), a targeted DNA methylation sequencing strategy. While RRBS provides sparse, biased coverage due to its enzymatic digestion and size-selection steps, meCUT&RUN delivers genome-wide signal with low sequencing requirements and base-pair resolution. In addition, meCUT&RUN captures 5mCs across a broad range of functional elements.
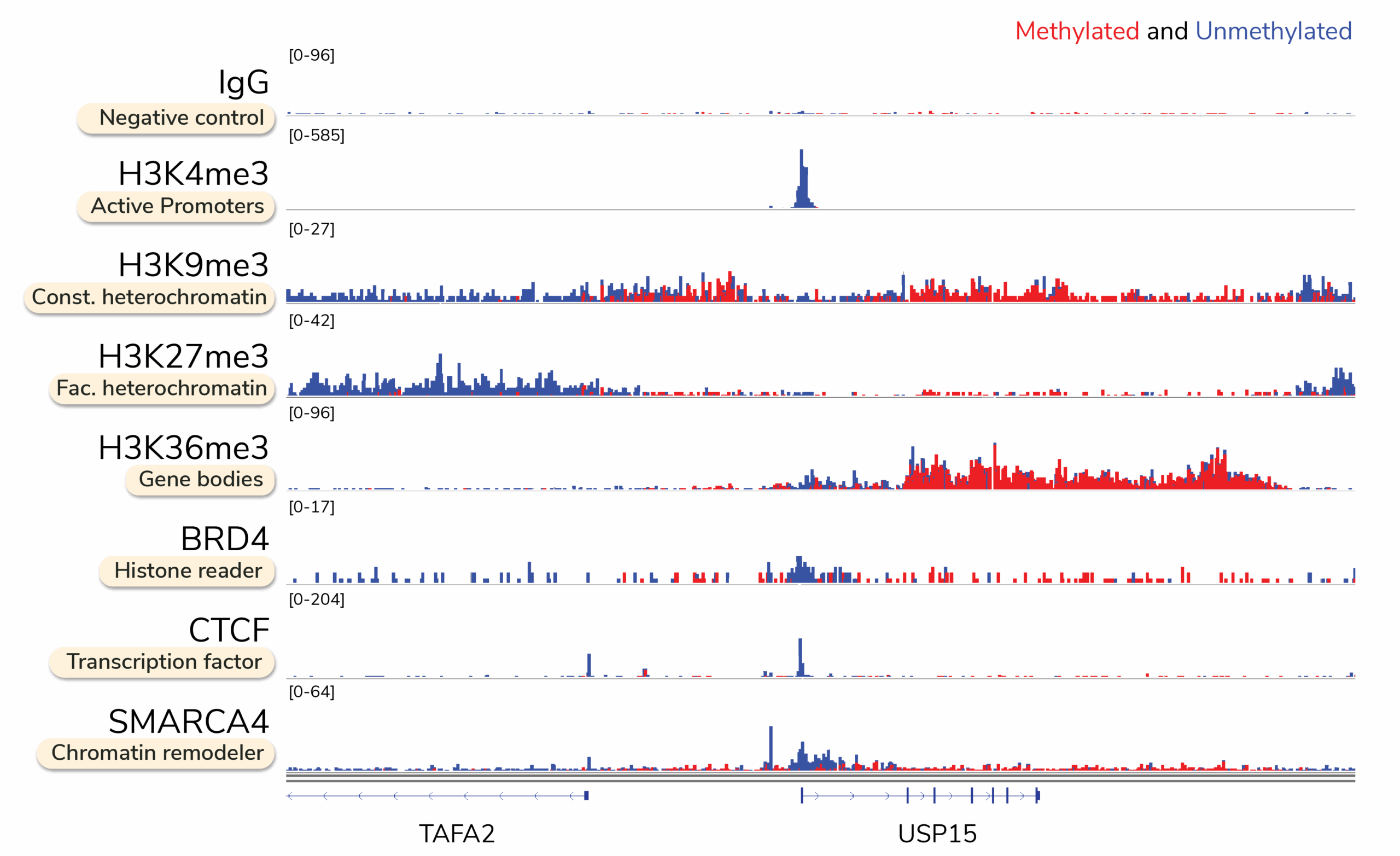
What does targeted DNA methylation mapping data look like with Multiomic CUT&RUN?
Traditional multiomic DNA methylation approaches combine chromatin immunoprecipitation sequencing (ChIP-seq) with whole-genome bisulfite sequencing (WGBS) datasets. However, these workflows are cost-prohibitive, require large numbers of cells, and take weeks (or longer) to complete.
Multiomic CUT&RUN streamlines this process by generating DNA methylation profiles specifically at protein-bound regions. This powerful approach produces high-quality genomic maps for transcription factor binding sites, histone post-translational modifications (PTMs), and chromatin-modifying enzymes, while simultaneously revealing differential DNA methylation patterns, such as hypermethylation at gene bodies and hypomethylation at promoters.
What does Fiber-seq data look like? What other insights does it provide?
Fiber-seq links genetic variants to epigenetic features in a single assay, providing a complete picture of chromatin architecture. The method uses a methyltransferase (Hia5) to label accessible adenines by methylating them, creating N6-methyladenine (6mA), effectively “stenciling” chromatin accessibility onto DNA. Next, long-read technologies such as Pacific Biosciences® (PacBio®) HiFi or Oxford Nanopore Technologies® (ONT) sequencing detect both the stenciled 6mA and endogenous DNA methylation, producing multiomic data along individual DNA molecules.
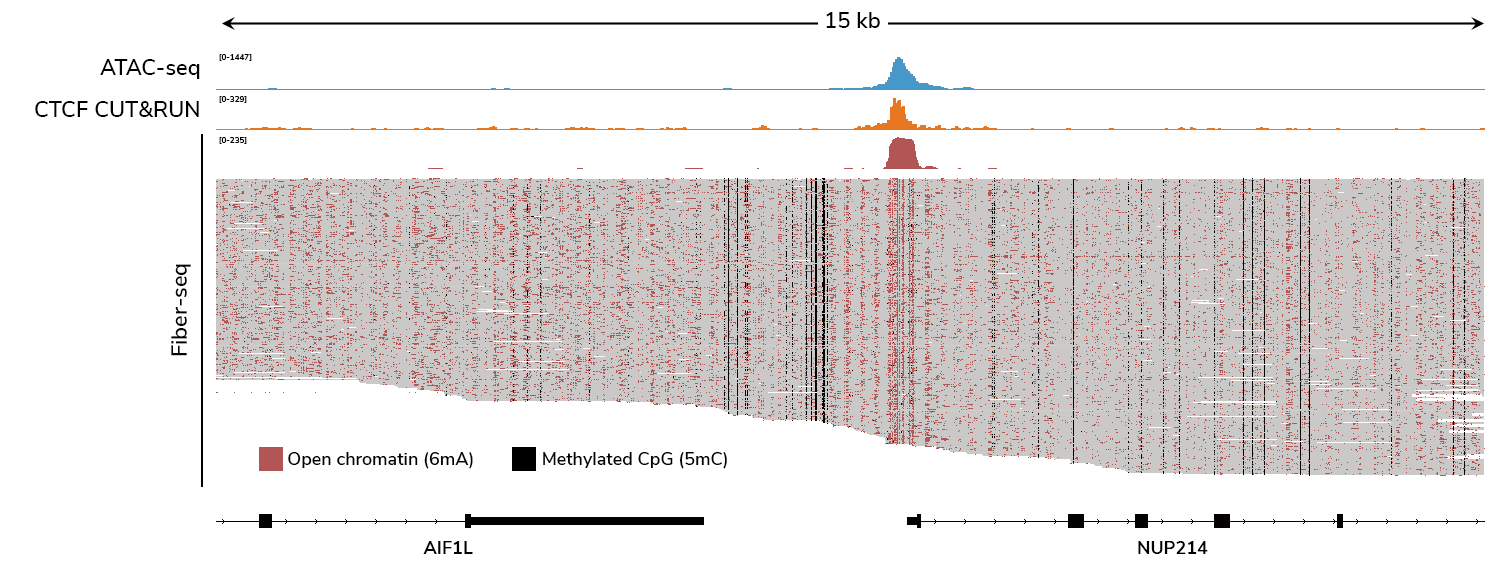
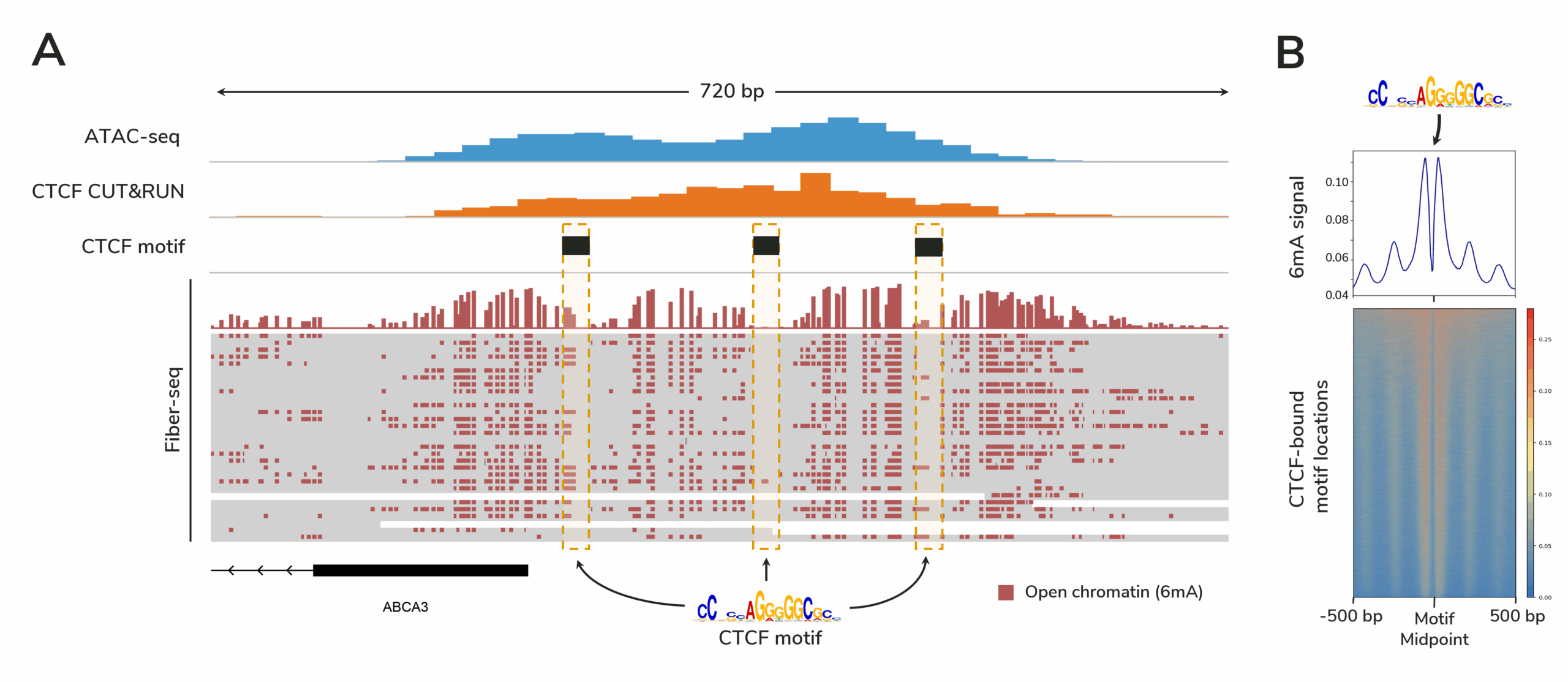
Because Fiber-seq directly profiles individual DNA molecules, it provides far higher resolution for detecting protein footprints than ATAC-seq—capturing transcription factors, nucleosomes, and even RNA polymerase II, all without the need for antibodies. Unlike ATAC-seq, Fiber-seq does not require averaging signals across a population of cells, revealing chromatin heterogeneity that bulk methods miss.
What tissues or cells types are compatible with our Genomic Services?
Our CUT&RUN Service protocols are optimized for freshly isolated and frozen cells or nuclei from various sources. For certain targets, such as labile histone lysine acetylation PTMs or acetyl-binding proteins, light cross-linking may be recommended. EpiCypher has successfully used our automated CUT&RUN assays for a variety of sample types and cell preparations, including:
- Suspension and adherent cell lines
- Drug-treated or stimulated cells
- Primary cells, including immune cells
- FACS-isolated cells
- Samples generated from tissue (including frozen patient biopsies)
- Clinical samples (PBMCs, BMMCs)
We provide detailed sample prep protocols and shipment instructions. For tissues, we can provide expert guidance on the best strategies for isolating cells.
We are actively developing genomic mapping assays compatible with formalin-fixed paraffin embedded (FFPE) tissues. Contact us to learn more about ongoing research efforts or to inquire for early access services that support this application.
For projects requiring long-read, single-molecule analysis, Fiber-seq offers broad sample compatibility and has been successfully applied to human cell lines, primary cells, plants, yeast, and non-model organisms. Reach out to our team to discuss whether Fiber-seq is the right fit for your project.
How many cells do I need for CUTANA Genomic Services?
For CUTANA CUT&RUN experiments, we recommend using 500,000 to 50,000 cells per reaction.*
Note that each reaction maps one target in one sample. Samples are unique cell populations, such as control vs. mutant or biological replicates.
If you have one cell sample, and you want to map three unique chromatin targets, you will be setting up three reactions – and thus require 1.5 million cells. We always suggest collecting 10-20% excess cells to account for sample loss.
* Our workflow is validated down to 10,000 cells for select targets. Success at low cell numbers depends on antibody performance, target abundance, and sample quality. There are additional risks assumed by going below the recommended cell numbers – such as low yields, increased adapter dimer contamination in libraries, increased background and/or sequencing depth – that may require additional optimization. Our scientists will help you weigh these risks during the experimental design phase of the project.
For Fiber-seq, input requirements differ because the assay relies on long-read sequencing and detection of Hia5-introduced 6mA. We recommend ~1,000,000 fresh nuclei per reaction, starting with ~2 million cells to ensure sufficient recovery after nuclei isolation. Flash-frozen and lightly cross-linked samples are also compatible, though labeling efficiency may vary slightly. Inputs below 1M nuclei are not yet validated. When working with non-human samples, keep in mind that genome size affects input needs—organisms with smaller genomes require proportionally more nuclei to match the total DNA content of 1 million human nuclei, ensuring robust labeling and sufficient DNA for long-read sequencing.
If you’re unsure how much material you’ll need, our team is happy to help you determine the optimal input for your specific sample type and assay.
What targets can I map? Can you help me find a good antibody?
The EpiCypher Services Team has experience mapping various targets:
- Transcription factors (e.g. CTCF, FOXA1, p53, c-Jun)
- Chromatin remodeling enzymes (e.g. SMARCA2/BRM and SMARCA4/BRG1)
- Histone modifying enzymes and regulators (e.g. MLL1, EZH2, BRD4, Menin)
- Histone PTMs (methylation, acetylation, phosphorylation, ubiquitination)
As a result, we have a broad catalog of target-specific antibodies demonstrating exceptional performance in CUT&RUN. We are also highly skilled in sourcing and validating antibodies to new targets. Antibody screening is offered as part of our Services and can be incorporated into your Statement of Work.
What data do I receive?
We work with every service partner to tailor our analysis to your needs. EpiCypher uses our bioinformatics pipeline to analyze sequencing data. At our data delivery meeting, we review preliminary data visualization at customer-identified target loci and all quality control checks (outlined above) to illustrate experimental success. We coordinate handoff of raw sequencing data, alignment files, bigWig files, and called peaks by SharePoint or Amazon Web Services. Files can be immediately used to visualize data, perform downstream analyses (e.g. differential enrichment), and derive biological insights in your lab.
To see a sample data report, email [email protected]