Scientific Posters
Last updated on April 5, 2024. Looking for a specific poster? Let us know here
Featured Posters
Recent Posters
| Poster Title | Presented At | Keywords | |
|---|---|---|---|
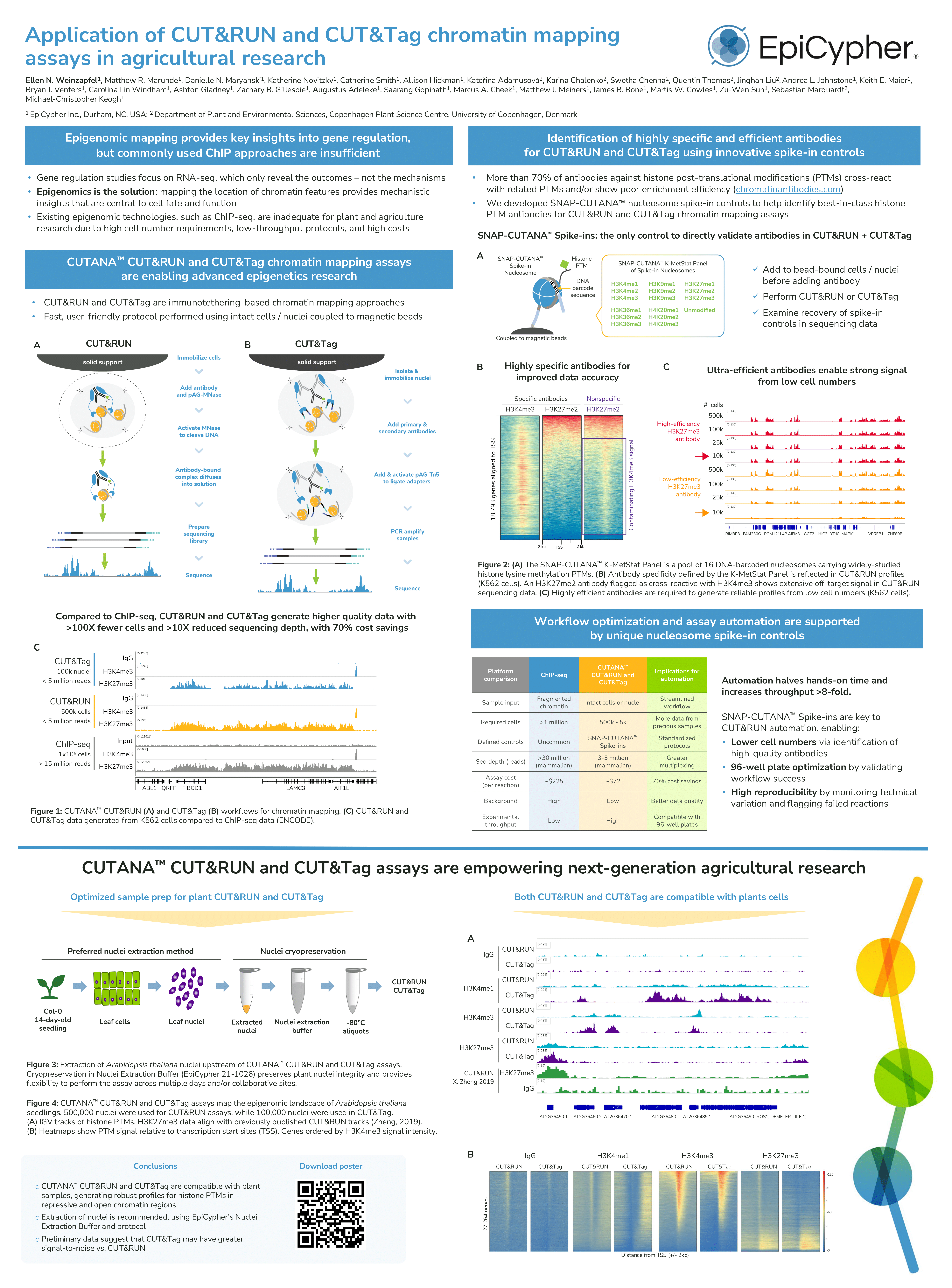
| CUT&RUN and CUT&Tag epigenomic mapping assays for agricultural research | AGBT Agriculture | CUT&RUN, CUT&Tag, agriculture | View Poster › |
| Exploring Antibodies and Rationally Designed Readers for Genomic Mapping of Histone Monoubiquitin | Keystone Epigenetic Mechanisms and Cancer Treatment | CUT&RUN, ubiquitination, antibody specificity | View Poster › |
| Development of a high-throughput CUT&RUN platform for epigenomic mapping of rare primary immune cells | Keystone Epigenetic Mechanisms and Cancer Treatment | Automated CUT&RUN, sorted cells, immune cells | View Poster › |
| Epigenomic fingerprinting of limited primary cells using automated CUT&RUN | AACR Blood Cancer Discovery | Automated CUT&RUN, sorted cells, immune cells | View Poster › |
| Direct multi-omics for the masses: Linking DNA methylation to chromatin targets via TEM-seq | AACR 2024 | DNA methylation, MeCP2, multiomic analysis | View Poster › |
| Multi-epigenomic mapping with long read sequencing | AACR 2024 | Multiomic analysis, long read sequencing, DNA methylation | View Poster › |
| CUTANA™ CUT&Tag : Powerful Platform for Streamlined, Ultra-Sensitive Epigenomics | Keystone Single-Cell Biology: Tissue Genomics, Technologies and Disease | CUT&Tag, CUTAC, chromatin accessibility | View Poster › |
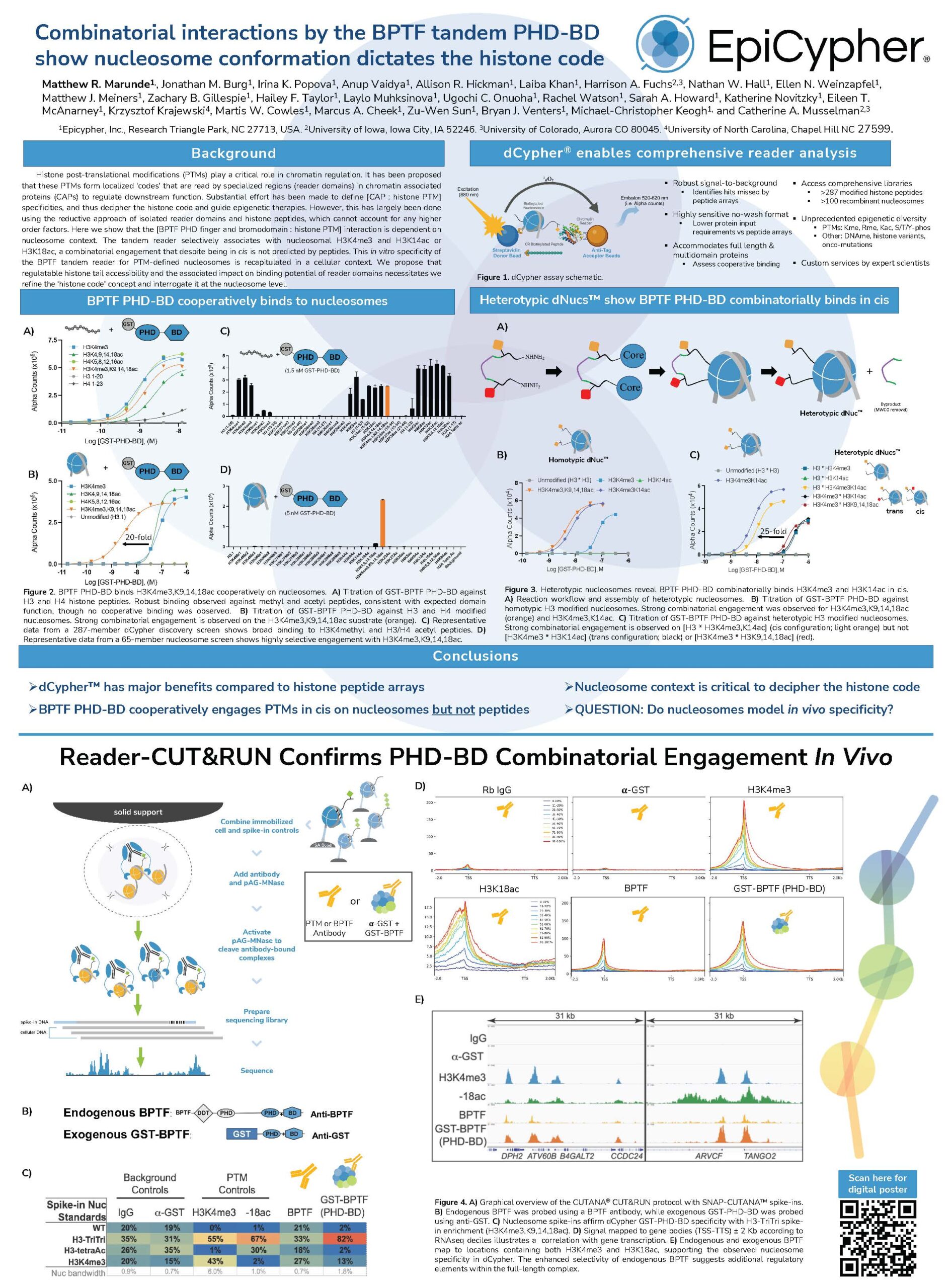
| Combinatorial interactions by the BPTF tandem PHD-BD show nucleosome conformation dictates the histone code | EpiCypher 2023 | Histone code, CUT&RUN, combinatorial histone PTMs | View Poster › |
| Development of Recombinant Reader Proteins for Epigenetic Profiling in Genomic Assays | EpiCypher 2023 | Reader CUT&RUN, antibody specificity, histone variants | View Poster › |
| Development of Quantitative Controls for CUT&RUN Assays | U.Wisconsin Epigenetics Symposium | CUT&RUN, spike-in controls, sequencing normalization | View Poster › |
| Automated CUT&RUN brings scalable epigenomic profiling to precision medicine | AGBT Precision Medicine | Automated CUT&RUN, DNA methylation, multiomic analysis | View Poster › |
| Nucleosome spike-in controls enable reliable next-generation epigenomic mapping | AACR Cancer Epigenomics | CUT&RUN, spike-in controls, sequencing normalization | View Poster › |
| dCypher™ screening of BPTF reader domains show nucleosomes, not peptides, dictate the histone code | Fusion Epigenetics | Histone code, CUT&RUN, combinatorial histone PTMs | View Poster › |
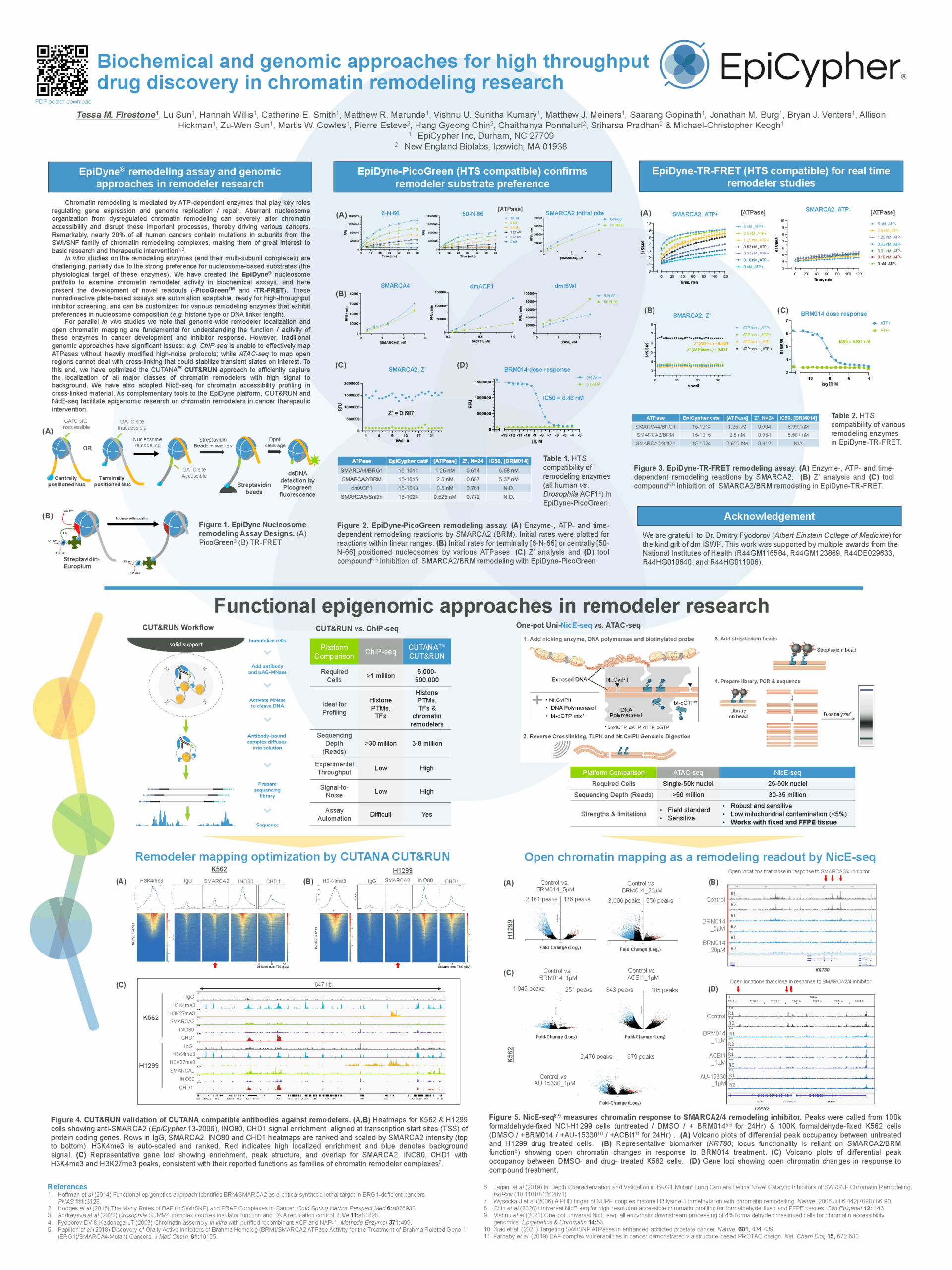
| Biochemical and genomic approaches for high throughput drug discovery in chromatin remodeling research | AACR 2023 | Chromatin remodeling, TR-FRET assays, CUT&RUN | View Poster › |
| Nucleosome-based screening in the context of epigenetic diversity enables improved methods to study histone modifications as disease biomarkers | EpiCypher 2021 | Histone Arginine Citrullination, Neutrophil Extracellular Trap (NETs), NETosis, Autoimmune Disease | View Poster › |
| Novel nucleosome-based methods for rapid screening and identification of best-in-class antibodies: a community resource to improve genomic mapping approaches | EpiCypher 2021 | ChIP-seq, Antibody Specificity, Luminex Screening, Nucleosome Spike-in Controls | View Poster › |
| High resolution epigenetics: Direct multiomics simultaneously maps DNA methylation and chromatin proteins | Keystone Precision Genome Engineering | DNA Methylation | View Poster › |
| CUT&RUN-EM: An Ultra-Sensitive Multiomic Method that Directly Links Chromatin Features to DNA Methylation | ASHG | DNA Methylation | View Poster › |
| Revolutionizing Epigenetics with Direct Multiomic Mapping of DNA Methylation and Chromatin Proteins | AGBT 2025 | CUT&RUN-EM, MeCUT&RUN | View Poster › |
| Development of a standards-driven genomic mapping platform to measure acute Pol II response dynamics | Max Planck Freiburg Epigenetics Meeting | RNA Pol II CUT&Tag, Transcriptional dynamics | View Poster › |
| Development of an ultrasensitive transcriptomic mapping platform to study opioid-regulated circuits in neuronal tissue | SfN | Transcription | View Poster › |
| Multiomic sequencing technology reveals crosstalk between chromatin proteins and DNA methylation in neurological disease | SfN | DNA methylation | View Poster › |
| Advanced multiomic sequencing technologies reveal complex crosstalk between chromatin proteins and DNA methylation in neurodevelopment | DOHaD | DNA methylation, multiomics, neurodevelopment, chromatin profiling | View Poster › |
| High resolution epigenetics: Direct multiomics simultaneously maps DNA methylation and chromatin proteins | Keystone Precision Genome Engineering | DNA methylation | View Poster › |
| Novel antibody development enables new insight into citrullination as a regulated modification | Keystone: Epigenetics in Development and Disease | View Poster › | |
| Utilizing RNA polymerase II to measure acute transcriptional changes | ASBMB Transcriptional Regulation | View Poster › | |
| Scaled Preparation of FFPE Tumor Samples for Epigenetic Studies | FASEB TCESRC | View Poster › | |
| Nucleosome spike-in controls enable reliable next-generation epigenomic mapping | AACR Cancer Epigenomics | CUT&RUN, spike-in controls, sequencing normalization | View Poster › |
| Multi-epigenomic mapping with long read sequencing | AGBT 2025 | Long Read Sequencing, chromatin profiling, methyltransferase, imprinting | View Poster › |