Chromatin Mapping Basics: ChIP-seq
- Nathaniel Wesley

Even if you are new to epigenetics, you have probably heard of ChIP-seq, CUT&RUN, and CUT&Tag – but how do these assays work? And why do scientists use them? Each of these methods are distinct chromatin mapping strategies, which scientists use to determine the location and enrichment of specific targets throughout the genome. A target can be a histone post-translational modification (PTM), a chromatin-associated protein (e.g. transcription factor), or even a covalent DNA modification (e.g. DNA methylation). The proper localization of these targets helps coordinate transcriptional programs in healthy cells/tissues, while dysregulation is considered a major contributor to human disease. As a result, understanding chromatin function is an important topic in biomedical and drug research.
ChIP (Chromatin ImmunoPrecipitation) has long been the gold standard technique for epigenomic mapping. Since the first ChIP experiments were published in the 1980s, more than 70,000 studies have used this technique or some variation1–3. In ChIP, crude fragmented chromatin is treated with target specific antibodies, which act to immunoprecipitate or “pull down” the target of interest and associated chromatin. DNA is purified from collected chromatin fragments, and then analyzed by next-generation sequencing (ChIP-seq) or qPCR (ChIP-qPCR). qPCR is used to determine target enrichment at a defined genomic location, while sequencing generates genome-wide localization profiles.
In this blog we will go through the general steps of ChIP, key optimization steps, and some important considerations before starting experiments. For further information and discussion on ChIP, we recommend these reviews4–7, our ChIP protocol chapter1, as well as our blog posts on chromatin mapping assays!
ChIP Basics
Step 1: Harvest and fix cells. The first step of a ChIP assay is harvesting cells. Typically, researchers will cross-link or “fix” cells using formaldehyde to stabilize association of chromatin-bound proteins.
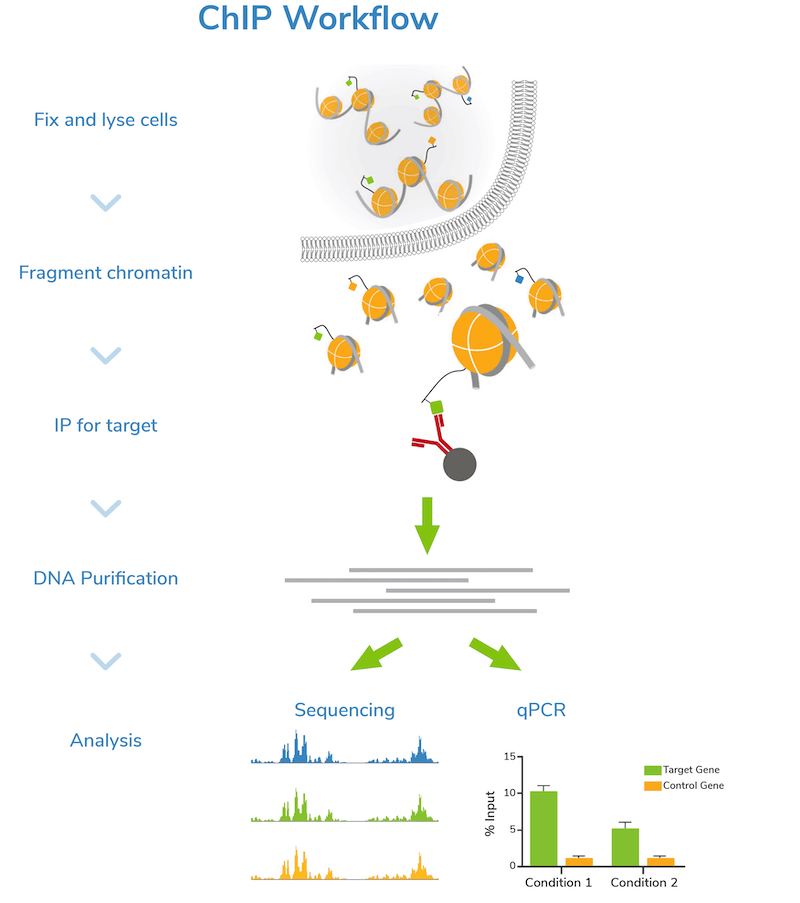
Step 2: Fragment chromatin. Next, cells are lysed, and bulk chromatin is isolated. The chromatin is fragmented or “sheared” to mononucleosome sized fragments (150-300 bp), which is important for obtaining high resolution sequencing data. Chromatin shearing is accomplished by sonication or enzymatic digestion and monitored by gel (e.g. agarose) or capillary electrophoresis (e.g. Agilent Bioanalyzer® or TapeStation®). An aliquot of fragmented chromatin is set aside as an “Input” control for quality control assessment and enrichment comparisons in later steps.
CRITICAL STEP: Chromatin shearing is the most challenging step of the ChIP workflow, requiring a delicate balance to achieve the desired level of fragmentation. See our notes on optimizing ChIP assays in the next section.
Step 3: Incubation with target-specific antibody. Sheared chromatin is incubated with a primary antibody, which binds to chromatin fragments carrying the target of interest. It is strongly recommended to include negative control reactions using an IgG antibody to assess background signal. If enough cells are available, using a validated positive control antibody (e.g. H3K4me3) is also suggested. Antibody incubation is typically performed overnight at 4˚C.
CRITICAL STEP: It is essential to use a high-quality antibody that efficiently captures its target with minimal cross-reactivity to other histone PTMs or chromatin-associated proteins. Read more about antibody selection below!
Step 4: Immunoprecipitation. The antibody is coupled to magnetic beads coated with protein-A and/or G (depending on the antibody isotype) to facilitate immunoprecipitation (Figure 1). Depending on the protocol, beads may be coupled to primary antibodies before the overnight antibody incubation, or on the following day. Regardless, the antibody-bound chromatin is isolated from bulk chromatin using a magnet, followed by a series of stringent washes. Wash buffers with progressively higher salt and detergent concentrations are used to strip away off-target proteins to reduce background signal in the data.
Step 5: DNA purification and quality check. The target-enriched chromatin is treated with Proteinase K to digest proteins, RNase A to degrade RNA, and high salt + heat to reverse cross-links. ChIP DNA is then purified using a DNA purification kit or equivalent method. DNA is also purified from the Input sample at this time.
The concentration of ChIP and Input DNA is assessed by spectrophotometric (e.g. Thermo Fisher Scientific Nanodrop™) or fluorometric (e.g. Invitrogen Qubit™) analysis. Agarose gel or capillary electrophoresis (Bioanalyzer® or TapeStation®) are used to confirm fragment size distribution prior to next steps. It is important to confirm that the ChIP DNA is enriched for mononucleosome-sized fragments, as opposed to very short or long pieces, to ensure successful downstream analysis.
Step 6: Next-generation sequencing (NGS) or qPCR. Now it’s time to analyze your sample! Sequencing allows genome-wide target studies, whereas qPCR provides a narrow look at specific region(s) of interest. The Input control is included in both strategies to determine enrichment. The IgG negative control is also analyzed to assess levels of nonspecific background signal.
For ChIP-seq studies, additional steps are required prior to sequencing. In brief:
Step 7: Library prep and indexing. ChIP DNA and Input DNA are repaired and amplified for NGS. During PCR, distinct indexes (or barcodes) are added to each unique library to allow multiplexed NGS. Multiplexing allows many samples to be sequenced in a single run.
Step 8: Library quantification and analysis. Prepared libraries are quantified and size distribution is confirmed, typically by capillary electrophoresis (Bioanalyzer® or TapeStation®).
Step 9: Sample pooling and loading onto the sequencer. For multiplexed sequencing, barcoded libraires are pooled at equimolar ratios and loaded onto the NGS platform. Time for NGS varies by platform, number of bases being sequenced, and whether paired-end sequencing is necessary.
The above steps are a simplified outline of a ChIP experiment. As any experienced chromatin scientist will tell you, ChIP is a challenging and time-consuming technique – there are multiple places for the assay to go awry.
What makes ChIP complicated? All the Optimization!
Every step of ChIP requires optimization. Furthermore, any modification to one step will likely affect downstream steps. For example, the number of cells required depends in part on your cross-linking conditions, the degree of chromatin fragmentation, as well as the specificity and enrichment efficiency of your antibody. Likewise, antibody selection impacts sequencing depth needed to achieve high signal over background. The interdependence of each step complicates assay development with iterative optimization quickly consuming significant time, money, and resources.
Below we highlight some common parameters to consider during ChIP optimization.
Number of Cells. The total number of cells needed depends on the intrinsic properties of your target, such as overall abundance, and efficacy of downstream processing steps. In general, ChIP reactions require a minimum of 500,000 cells; but in practice, most researchers use million(s) of cells per ChIP.
Replicates. Replicates are also important for ChIP, since the data can vary across reactions. You should plan for three biological replicates per target. A biological replicate represents independently collected cell or tissue samples. For instance, if you want to map H3K4me3 in intestinal epithelial cells from control mice and mutant mice models, you will want to harvest tissue from three controls and three mutant mice. Conversely, a technical replicate represents a repeated procedural step performed on the same biological sample (e.g. ChIP replicates performed using Input DNA from the same mouse)8. Technical replicates may be considered in early optimization stages to ensure robust conditions for future experiments.
Cross-linking conditions. This step requires careful optimization for your cell type and the target of interest. Parameters to adjust include the number of cells harvested as well as the cross-linking agent (e.g. formaldehyde, glutaraldehyde, disuccinimidyl glutarate [DSG]), agent concentration, and incubation time. Excessive cross-linking can mask the antibody epitope, prevent effective chromatin shearing, and degrade the sample. Alternatively, insufficient cross-linking can increase sensitivity to mechanical shearing processes, reduce total captured target chromatin, and impact data analysis. Typically, a time-course experiment is performed to determine the optimal cross-linking time and concentration.
Note on Native ChIP: For highly stable histone-DNA interactions, or for targets tightly associated with DNA (e.g. histone PTMs), cross-linking is not necessary and ChIP can be performed under native, unfixed conditions. However, in native ChIP workflows, the nuclei are first isolated from cells before fragmentation. See EpiCypher’s recently published protocol chapter for detailed native ChIP methods1.
Chromatin fragmentation. Chromatin shearing is routinely highlighted as one of the most difficult parts of ChIP experiments. Generally, a size range from 150-300 bp is desirable (i.e. between mononucleosome and dinucleosome fragments). If the pieces of chromatin are too long (>600-700 bp), it becomes difficult to identify exactly where the protein or histone PTM is located, thus lowering resolution. Furthermore, larger fragments immunoprecipitate more efficiently due to antibody avidity bias9. Large DNA fragments are also unsuitable for most next generation sequencing (NGS) platforms, which prefer genomic DNA fragment sizes of 200-600 bp. On the other hand, excessive fragmentation disrupts target interactions and reduces ChIP yields.
Fragmentation methods: In standard cross-linked ChIP workflows, bulk chromatin can be fragmented by sonication or enzymatic digestion using micrococcal nuclease (MNase). Native ChIP samples require MNase digestion, as mechanical shearing processes can disrupt unfixed chromatin and alter genomic profiles.
Optimization: For both sonication and digestion, a time course is performed to determine the ideal number of sonication cycles/digestion time, ideal ratio of buffer volume vs. number of cells, and compatibility with cross-linking conditions. It is recommended to optimize fragmentation conditions for every new input type (e.g. new cell/tissue type, varied treatment/processing steps), since cross-linking conditions and cell viscosity impact fragmentation. Even for small protocol changes, it is a good idea to check that fragmentation conditions are working as expected.
Antibody selection. The success of a ChIP experiment is dependent on using a highly specific and efficient antibody. This means that the antibody shows low binding to off-target proteins or histone PTMs, and that the antibody recovers a high percentage of the target. How should antibodies be selected?
Histone PTM antibodies notoriously show high levels of cross-reactivity in ChIP which can mislead biological conclusions1,9–13. To address this issue, EpiCypher developed SNAP-ChIP® Spike-in technology, which uses DNA-barcoded designer nucleosomes to assess histone PTM antibody performance directly in a ChIP experiment. This approach is more reliable than surrogate assays (e.g. histone peptide arrays, immunoblot, ELISA) that do not adequately mimic ChIP10. We also offer a collection of SNAP-ChIP Certified Antibodies to histone PTM targets, which have been extensively validated for high specificity and efficiency in ChIP assays. Be sure to visit chromatinantibodies.com and look up your histone PTM for detailed information on commercial antibody efficiency and specificity.
If trying to identify antibodies for chromatin-associated proteins, or if no ChIP-grade antibodies are available, it is recommended to source 3-5 antibodies from various reputable vendors and that bind distinct epitopes. Do a side-by-side comparison and select the best antibody based on ChIP yields, enrichment, and generation of expected peak structures consistent with the target function(s).
Note on lot-specific testing: Antibody performance can vary drastically lot-to-lot, even for some monoclonal antibodies. EpiCypher recommends re-testing every new lot of antibody to confirm specificity and efficiency is maintained in ChIP. If you find an excellent antibody to a target of interest, it is recommended to stock up on aliquots from the same production lot!
Immunoprecipitation (IP). The IP step is inherently noisy. In addition to problems with antibody performance, the magnetic beads may pull down off-target fragments along with the chromatin containing the target. IP also requires highly stringent washes, which can reduce yields and lower signal-to-noise.
Optimization: Test different antibodies, antibody concentrations, IP incubation temperatures and times. For some low abundance targets, yields can be improved by increasing the number of cells. In addition, both over and under cross-linking and insufficient chromatin fragmentation influence IP.
Impact on sequencing and qPCR. Even the most well-optimized ChIP assays can show high background, which increases experimental variation and complicates data interpretation. ChIP-seq requires 20 million or more sequencing reads per reaction to detect peak enrichment over background, driving up costs and consuming resources.
Is There an Alternative to ChIP-seq?
Despite extensive optimization, millions of cells, and high sequencing depths, ChIP-seq remains a highly variable assay and is not guaranteed to work. Luckily there are exciting new technologies that are making massive steps forward in chromatin profiling, such as CUT&RUN and CUT&Tag.
CUT&RUN and CUT&Tag assays use fusion enzymes to selectively target antibody-bound chromatin in intact cells. These strategies address or eliminate many pain points associated with ChIP, outlined in Figure 2.

But how do these assays work? Are they ideal for all kinds of targets and cell types? We will be back soon to discuss more! In the meantime, if you are starting CUT&RUN or CUT&Tag, check out this blog for guidance. We also offer a collection of CUTANA™ CUT&RUN and CUT&Tag kits, enzymes, antibodies, and accessory reagents, as well as robust protocols validated for diverse targets.
References
- Small, E. C. et al. Chromatin Immunoprecipitation (ChIP) to Study DNA–Protein Interactions. Methods Mol. Biol. 2261, 323–343 (2021). PubMed PMID: 33420999.
- Gilmour, D. S., and Lis, J. T. Detecting protein-DNA interactions in vivo: Distribution of RNA polymerase on specific bacterial genes. Proc. Natl. Acad. Sci. U. S. A. 81, 4275–4279 (1984). PubMed PMID: 3018544.
- Gilmour, D. S., and Lis, J. T. In vivo interactions of RNA polymerase II with genes of Drosophila melanogaster. Mol. Cell. Biol. 5, 2009–2018 (1985). PubMed PMID: 3018544.
- Park, P. J. ChIP-seq: Advantages and challenges of a maturing technology. Nat. Rev. Genet. 10, 669–680 (2009). PubMed PMID: 19736561.
- Furey, T. S. ChIP-seq and beyond: New and improved methodologies to detect and characterize protein-DNA interactions. Nat. Rev. Genet. 13, 840–852 (2012). PubMed PMID: 23090257.
- Farnham, P. J. Insights from genomic profiling of transcription factors. Nat. Rev. Genet. 10, 605–616 (2009). PubMed PMID: 19668247.
- Landt, S. G. et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 22, 1813–1831 (2012). PubMed PMID: 22955991.
- Blainey, P. et al. Points of significance: Replication. Nat. Methods 11, 879–880 (2014). PubMed PMID: 25317452.
- Grzybowski, A. T. et al. Calibrating ChIP-Seq with Nucleosomal Internal Standards to Measure Histone Modification Density Genome Wide. Mol. Cell 58, 886–899 (2015). PubMed PMID: 26004229.
- Shah, R. N. et al. Examining the Roles of H3K4 Methylation States with Systematically Characterized Antibodies. Mol. Cell 72, 162-177.e7 (2018) PubMed PMID: 30244833.
- Fuchs, S. M. et al. Influence of combinatorial histone modifications on antibody and effector protein recognition. Curr. Biol. 21, 53–58 (2011). PubMed PMID: 21167713.
- Fuchs, S. M., and Strahl, B. D. Antibody recognition of histone post-translational modifications: Emerging issues and future prospects. Epigenomics 3, 247–249 (2011). PubMed PMID: 2212232.
- Baker, M. Blame it on the antibodies. Nature 521, 274–276 (2009). PubMed PMID: 25993940.
