CUTANA™ DNA Purification Beads
CUTANA™ DNA Purification Beads harness Solid-Phase Reversible Immobilization (SPRI*) technology to deliver fast, high-yield DNA cleanup and precise size selection for NGS library preparation and other genomic workflows. Optimized PEG and salt concentrations drive efficient, reproducible binding of DNA to paramagnetic beads, ensuring exceptional recovery with minimal bias or carry-over contamination. The formulation matches the bead-to-sample ratio of popular alternatives such as AMPure XP* and SPRIselect*, ensuring CUTANA™ DNA Purification Beads can serve as a drop-in replacement into manual or automated cleanup workflows without any protocol re-optimization.
Key Advantages
- Trusted method – SPRI chemistry researchers rely on for DNA cleanup
- NGS validated – proven performance in genomic workflows including CUT&RUN, CUT&Tag, and library preparation
- High recovery & purity – consistently recovers DNA across a wide range of fragments and input amounts
- Automation-compatible – suitable for manual or automated protocols using magnetic separation
- Drop-in AMPure XP* alternative – identical ratios for seamless replacement in SPRI-based cleanups
- Room-temperature stable – ready to use; no warmup period required
Validated Applications
- CUT&RUN
- CUT&Tag
NGS Library Preparation & adapter dimer cleanup
*SPRI, SPRIselect, and AMPureXP are registered trademarks of Beckman Coulter and nominative references to them are for descriptive purposes only. CUTANA™ products are not manufactured by or affiliated with Beckman Coulter.
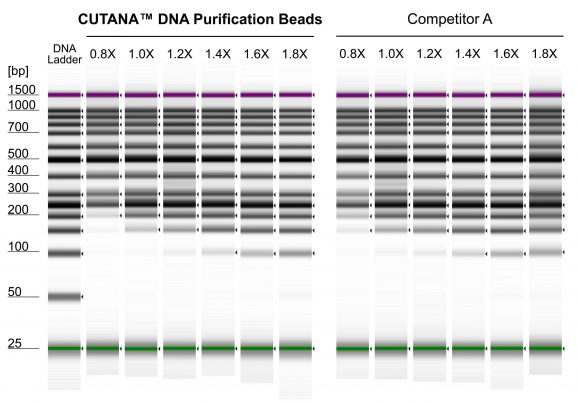
Figure 1: DNA Fragment Recovery Cleanup
Representative gel images show recovery of various DNA fragment sizes generated using CUTANA™ DNA Purification beads compared to Competitor A. CUTANA™ DNA Purification Beads were subjected to a magnetic bead-based DNA cleanup protocol using a GeneRuler 50 bp DNA Ladder (ThermoFisher Scientific SM0371) tested at a range of bead-to-sample ratios (0.8 – 1.8X). The purified DNA was run on an Agilent TapeStation.
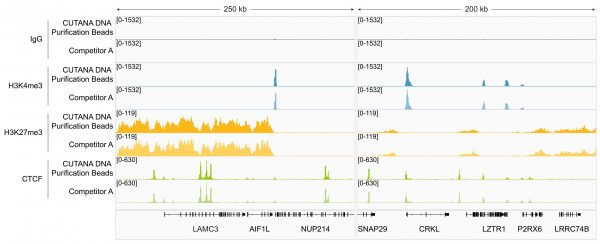
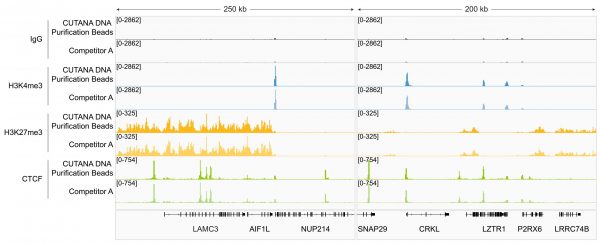
Figure 2: Magnetic Bead-based DNA Cleanup in CUT&RUN
CUTANA™ DNA Purification Beads perform equivalently to Competitor A beads in CUT&RUN. CUT&RUN was performed on 500k K562 cells using the CUTANA™ ChIC/CUT&RUN Kit v5 (EpiCypher 14-1048) with 0.5 µg of either IgG negative control (EpiCypher 13-0042), H3K4me3 positive control (EpiCypher 13-0060), H3K27me3 positive control (EpiCypher 13-0055), or CTCF (EpiCypher 13-2014) antibodies. CUTANA™ DNA Purification Beads (see Application Notes) or Competitor A beads were used for CUT&RUN DNA purification. Library preparation was performed with 5 ng of DNA (or the total amount recovered if less than 5 ng) using the CUTANA™ CUT&RUN Library Prep Kit (EpiCypher 14-1001/14-1002). Libraries were run on an Illumina NextSeq2000 with paired-end sequencing (2×50 bp). Sample sequencing depth was 12.5/15.1 million reads (IgG, CUTANA/Competitor A), 14.1/18.8 million reads (H3K4me3, CUTANA/Competitor A), 16.4/13.5 million reads (H3K27me3, CUTANA/Competitor A), and 14.0/14.1 million reads (CTCF, CUTANA/Competitor A). Data were aligned to the hg38 genome using Bowtie2. Data were filtered to remove duplicates, multi-aligned reads, and ENCODE DAC Exclusion List regions. Gene browser shots were generated using the Integrative Genomics Viewer (IGV, Broad Institute). Two representative loci are shown.
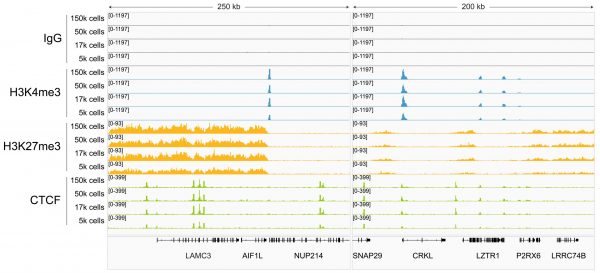
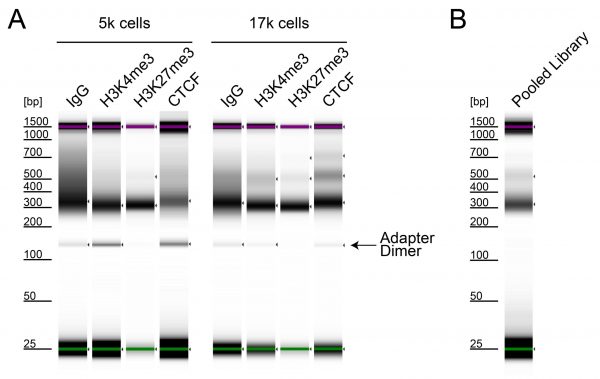
Figure 3: Magnetic Bead-based DNA Cleanup in CUT&RUN at Low Cell Inputs
CUTANA™ DNA Purification Beads perform robustly in CUT&RUN, even at low cell numbers. CUT&RUN was performed on 150k, 50k, 17k, or 5k K562 cells using the CUTANA™ ChIC/CUT&RUN Kit v5 (EpiCypher 14-1048) with 0.5 µg of either IgG negative control (EpiCypher 13-0042), H3K4me3 positive control (EpiCypher 13-0060), H3K27me3 positive control (EpiCypher 13-0055), or CTCF (EpiCypher 13-2014) antibodies. CUTANA™ DNA Purification Beads were used for DNA purification (see Application Notes). Library preparation was performed with 5 ng of DNA (or the total amount recovered if less than 5 ng) using the CUTANA™ CUT&RUN Library Prep Kit (EpiCypher 14-1001/14-1002). Libraries were run on an Illumina NextSeq2000 with paired-end sequencing (2×50 bp). Sample sequencing depth was 7.2/6.3/5.5/6.6 million reads (IgG, 150k/50k/17k/5k cell input), 6.8/6.6/5.6/6.6 million reads (H3K4me3, 150k/50k/17k/5k cell input), 8.9/6.0/7.5/7.1 million reads (H3K27me3, 150k/50k/17k/5k cell input), and 4.8/5.6/5.5/5.8 million reads (CTCF, 150k/50k/17k/5k cell input). Data were aligned to the hg38 genome using Bowtie2. Data were filtered to remove duplicates, multi-aligned reads, and ENCODE DAC Exclusion List regions. Gene browser shots were generated using the Integrative Genomics Viewer (IGV, Broad Institute). Two representative loci are shown.
Figure 4: Magnetic Bead-based DNA Cleanup in CUT&Tag
CUTANA™ DNA Purification Beads perform equivalently to Competitor A beads in CUT&Tag. CUT&Tag was performed on 100k K562 nuclei using the CUTANA™ CUT&Tag Kit v3 (EpiCypher 14-1102/14-1103) with 0.5 µg of either IgG negative control (EpiCypher 13-0042), H3K4me3 positive control (EpiCypher 13-0060), H3K27me3 positive control (EpiCypher 13-0055), or CTCF (EpiCypher 13-2014) antibodies. CUTANA™ DNA Purification Beads (see Application Notes) or Competitor A beads were used for PCR cleanup. Libraries were run on an Illumina NextSeq2000 with paired-end sequencing (2×50 bp). Sample sequencing depth was 15.6/13.1 million reads (IgG, CUTANA/Competitor A), 12.1/14.4 million reads (H3K4me3, CUTANA/Competitor A), 18.2/14.3 million reads (H3K27me3, CUTANA/Competitor A), and 13.7/13.6 million reads (CTCF, CUTANA/Competitor A). Data were aligned to the hg38 genome using Bowtie2. Data were filtered to remove duplicates, multi-aligned reads, and ENCODE DAC Exclusion List regions. Gene browser shots were generated using the Integrative Genomics Viewer (IGV, Broad Institute). Two representative loci are shown.
Figure 5: Magnetic Bead-based Cleanup of Adapter Dimers from Genomic Sequencing Libraries
Adapter dimers (~125 bp) result from self-ligation of sequencing adapters that may be preferentially amplified in PCR. They are particularly problematic at low cell inputs, where genomic DNA templates are scarce. Adapter dimers occupy valuable sequencing bandwidth and should be removed whenever possible. Agilent TapeStation revealed adapter dimers present in CUT&RUN libraries generated using 5k and 17k K562 cells (A). CUTANA™ DNA Purification Beads were used for DNA size selection (see Application Notes) on the pooled sequencing library, resulting in high-quality ready-for-sequencing libraries (B).
Paramagnetic bead suspension
Store at 15 – 30°C (room temperature; DO NOT FREEZE). Stable for 12 months from date of receipt.
Binding Mechanism
SPRI (PEG/NaCl)
Tunable via 0.8X – 1.8X bead-to-sample ratio
HandlingUse CUTANA™ DNA Purification Beads at the following bead-to-sample ratios:
DNA Purification: 1.8X
CUT&TagPCR Cleanup: 1.3X
Adapter Dimer Removal: 1.0X
Total DNA Cleanup: 1.8X