CUTANA™ CUT&Tag Kit
The CUTANA™ CUT&Tag Kits you know and trust, now with NEW catalog numbers!
Primer Set 1
OLD Cat No: 14-1102 → NEW Cat No: 14-1102-48s1
Primer Set 2
OLD Cat No: 14-1103 → NEW Cat No: 14-1102-48s2
NEW! 24-Reaction Kit with Primer Set 1 Subset
Cat No: 14-1102-24s3
The CUTANA™ CUT&Tag Kit offers a comprehensive solution for ultra-sensitive mapping of histone post-translational modifications (PTMs). Key advantages include:
- All-inclusive solution – The CUTANA™ CUT&Tag Kit includes all reagents needed to go from cells to sequencing libraries (including sequencing primers!).
- User-friendly workflow – Streamlined 8-strip tube workflow and exclusive direct-to-PCR protocol ensures increased throughput and minimal sample loss.
- Controls drive reliability – Control antibodies and nucleosome spike-ins enable assay optimization and continuous monitoring.
- Flexible pack sizes for every workflow – Support diverse research needs from first-time users to high-throughput studies by multiplexing up to 96 reactions.
- Extensive support library – The kit manual and Tech Support Center provide comprehensive guides for experimental design, quality checks, and FAQs.
Stringent quality control – Each new lot undergoes rigorous quality control checks to ensure consistent results in your research.
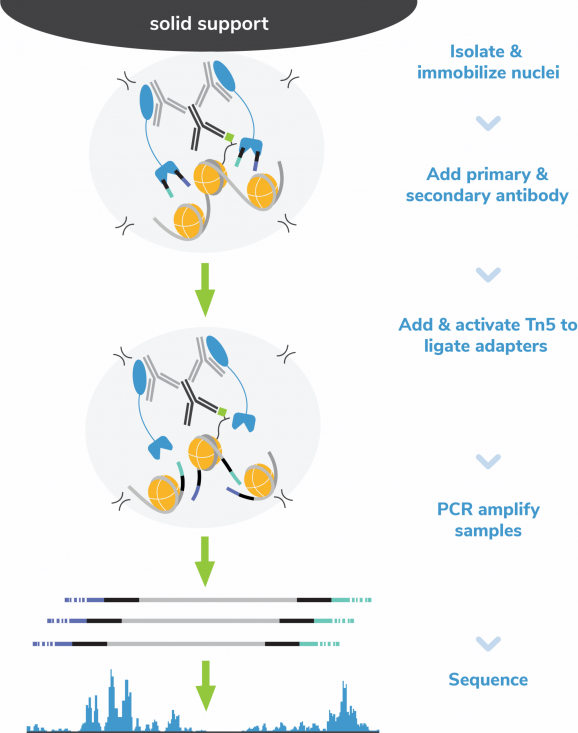
In CUT&Tag, antibody-bound chromatin is selectively cleaved with fusion protein pAG-Tn5, which simultaneously adds sequencing adapters. Tagmented fragments bypass traditional library prep with CUTANA™ CUT&Tag Kit’s exclusive Direct-to-PCR strategy, enabling users to go from cells to PCR-amplified sequencing libraries in one tube with minimal sample loss. The recommended input for CUT&Tag is 100,000 native nuclei per reaction. Comparable data can be generated down to 10,000 nuclei, and the protocol is also validated for whole cells, cryopreserved samples, and lightly cross-linked nuclei or cells. CUT&Tag provides robust profiling for histone PTMs; for chromatin-associated proteins (e.g. transcription factors), CUTANA™ CUT&RUN is recommended (EpiCypher 14-1048, EpiCypher 14-1001).
The CUT&Tag Kit Version 4 (v4) now includes two new Wash Buffer Enhancers, CUTANA™ DNA Purification beads, and improvements to the CUT&Tag protocol. CUTANA™ DNA Purification Beads are optimized for high yield DNA clean up with precise size selection, while the CUTANA™ Wash Buffer Enhancers reduce clumping and improve bead handling. Protocol improvements amplify sample preservation with changes to nuclei resuspension and refine reaction handling to increase yields for difficult targets. The protocol is also designed for compatibility with multi-channel pipetting for increased throughput and reproducibility. Positive (H3K4me3 and H3K27me3) and negative (IgG) control antibodies are paired with the SNAP-CUTANA™ K-MetStat Panel of nucleosome spike-in controls (Figure 2) to continuously monitor workflows and guide troubleshooting. CUTANA™ CUT&Tag Kits are bench-tested, scientist-approved, providing users with quality reagents for precision mapping.
Intellectual Property
US Pat. No. 10689643, 11306307, 11733248, 10732158, 10087485; EU Pat. No. 3688157, 2999784, 3102721, 2859139; JP Pat. No. 6985010, 6293742; CN Pat. No. 2859139 and related patents and applications.
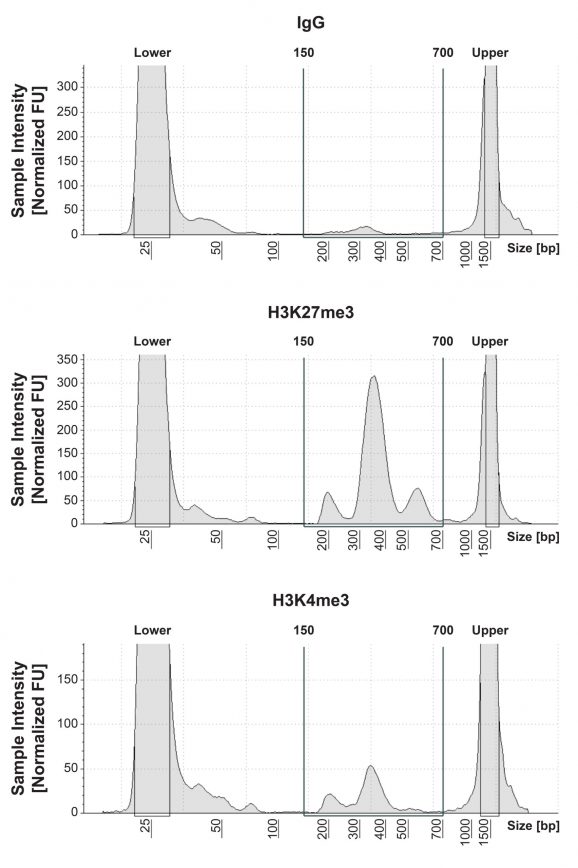
Figure 1: CUT&Tag DNA fragment size distribution analysis
CUT&Tag was performed as described in Figure 5. Library DNA was analyzed by Agilent TapeStation®, which confirmed that mononucleosomes were predominantly enriched in CUT&Tag (peak between 300-400 bp). Peak between 500-700 bp represents dinucleosomes.
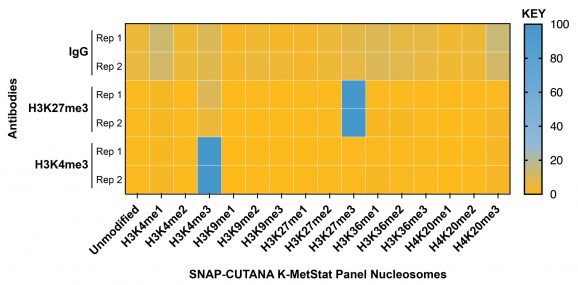
Figure 2: SNAP-CUTANA™ K-MetStat Spike-in controls
DNA-barcoded designer nucleosomes (dNucs) representing 16 different K-methyl PTM states: mono-, di-, and tri-methylation at H3K4, H3K9, H3K27, H3K36, and H4K20, as well as unmodified control, were spiked into CUT&Tag samples prior to the addition of the control antibodies provided with the kit (IgG, H3K27me3, H3K4me3). After sequencing, instances of each spike-in barcode recovered in the CUT&Tag reactions were counted and normalized from raw fastq files using the shell script and analysis Excel sheet available on the spike-in product page (EpiCypher 19-1002). Barcodes for IgG (top; normalized to the sum of total reads), H3K27me3 (middle; normalized to on-target), and H3K4me3 (bottom; normalized to on-target) antibodies provided with this kit are shown. The spike-ins confirmed optimal experimental conditions (H3K27me3 and H3K4me3 antibodies specifically recovered the target dNuc, while IgG showed no preferential enrichment).
Figure 3: CUT&Tag genome-wide heatmaps
CUT&Tag was performed as described in Figure 5. Heatmaps show two replicates (“Rep”) of IgG and H3K4me3 antibodies in aligned rows ranked by intensity (top to bottom) relative to the H3K4me3 Rep 1 reaction. High, medium, and low intensity are shown in red, yellow, and blue, respectively. Antibodies to histone PTMs showed expected enrichment patterns and high reproducibility. H3K4me3, a marker of active transcription localized to transcription start sites (TSSs), shows enrichment consistent at TSSs, as expected. IgG shows low background enrichment.
Figure 4: Representative gene browser tracks
CUT&Tag was performed as described in Figure 5. A representative 186 kb window at the LAMC3 gene is shown for two replicates (“Rep”) of IgG, H3K27me3, and H3K4me3 kit control antibodies. Representative tracks are also shown for two replicates of H3K4me1 antibody. The CUT&Tag kit produced the expected genomic distribution for each target. Images were generated using the Integrative Genomics Viewer (IGV, Broad Institute).
Figure 5: CUT&Tag methods
CUT&Tag was performed using the CUTANA™ CUT&Tag Kit starting with 100k K562 cells and 0.5 µg of either IgG (EpiCypher 13-0042t), H3K27me3 (EpiCypher 13-0055t), H3K4me3 (EpiCypher 13-0060t), or H3K4me1 (EpiCypher 13-0057) antibodies. Libraries were run on an Illumina NextSeq2000 with paired-end sequencing (2×50 bp). Sample sequencing depth was 5.3/4.1 million reads (IgG Rep 1/Rep 2), 11.2/9.0 million reads (H3K27me3 Rep 1/Rep 2), 8.6/5.0 million reads (H3K4me3 Rep 1/Rep 2), and 4.1/10.3 million reads (H3K4me1 Rep 1/Rep 2). Data were aligned to the hg19 genome using Bowtie2. Data were filtered to remove duplicates, multi-aligned reads, and ENCODE DAC Exclusion List regions.
Storage
OPEN KIT IMMEDIATELY and store components at room temperature, 4°C, and -20°C as indicated (see Kit Manual). Stable for 12 months upon date of receipt.
Instructions for Use
See Kit Manual
| Item | Cat. No. |
|---|---|
| CUTANA™ pAG-Tn5 for CUT&Tag | 15-1017t |
| H3K27me3 Antibody, SNAP-Certified™ for CUT&RUN and CUT&Tag | 13-0055t |
| H3K4me3 Antibody, SNAP-Certified™ for CUT&RUN and CUT&Tag | 13-0060t |
| CUTANA™ Rabbit IgG CUT&RUN Negative Control Antibody | 13-0042t |
| Anti-Rabbit Secondary Antibody for CUTANA™ CUT&Tag Workflows | 13-0047t |
| SNAP-CUTANA™ K-MetStat Panel | 19-1002t |
| CUTANA™ Concanavalin A Conjugated Paramagnetic Beads | 21-1401t |
| CUTANA™ Non-Hot Start 2X PCR Master Mix for CUT&Tag | 15-1018t |
| CUTANA™ CUT&RUN 8-strip 0.2 mL Tubes | 10-0009t |
| 5.0 M NaCl | 21-1013t |
| 0.5 M EDTA | 21-1006t |
| 1 M MgCl2 | 21-1015t |
| SDS Release Buffer | 21-1017t |
| SDS Quench Buffer | 21-1018t |
| 0.1X TE Buffer | 21-1025t |
| Pre-Wash Buffer | 21-1002t |
| Pre-Nuclei Extraction Buffer | 21-1021t |
| Bead Activation Buffer | 21-1001t |
| 5% Digitonin | 21-1004t |
| 1 M Spermidine | 21-1005t |
| Wash Buffer Enhancer 1 | 21-1028t |
| Wash Buffer Enhancer 2 | 15-1030t |
| CUTANA™ DNA Purification Beads | 21-1407t |
| Multiplexing Primers | 14-1102-48s1 and 14-1102-48s2 each contain 14-1102-24s3 includes a subset of Primer Set 1 and |
| Item | Cat. No. |
|---|---|
| Anti-Mouse Secondary Antibody for CUTANA™ CUT&Tag Workflows | 13-0048 |
| SNAP-CUTANA™ K-MetStat Panel | 19-1002 |
| CUT&Tag Antibodies | See the list |
| Magnetic Separation Rack, 0.2 mL Tubes | 10-0008 |
| Magnetic Separation Rack, 1.5 mL Tubes | 10-0012 |
| CUTANA™ Protease Inhibitor Tablets | 21-1027 |
| CUTANA™ Quick Cleanup DNA Purification Kit | 14-0052 |
Older Versions |