BRG1/SMARCA4 CUTANA™ CUT&RUN Antibody
In stock
This antibody meets EpiCypher’s “CUTANA Compatible” criteria for performance in Cleavage Under Targets and Release Using Nuclease (CUT&RUN) and/or Cleavage Under Targets and Tagmentation (CUT&Tag) approaches to genomic mapping. Every lot of a CUTANA Compatible antibody is tested in the indicated CUTANA approach using EpiCypher optimized protocols and determined to yield peaks that show a genomic distribution pattern consistent with reported function(s) of the target protein. BRG1 antibody produces CUT&RUN peaks primarily flanking transcription start sites (TSSs, Figure 1). BRG1 peaks show a large degree of overlap with BRD4 peaks (Figure 2), as has been reported in the literature [Conrad et al., 2017].
Trademarks and copyrights
CUT&RUN was performed on 500k K562 cells with 0.5 µg of either BRG1/SMARCA4, H3K4me3 positive control (EpiCypher 13-0041), or IgG negative control (EpiCypher 13-0042) antibodies using the CUTANA™ ChIC/CUT&RUN Kit v2.0 (EpiCypher 14-1048). Library preparation was performed with 5 ng of DNA (or the total amount recovered if less than 5 ng) using the CUTANA™ CUT&RUN Library Prep Kit (EpiCypher 14-1001/14-1002). Both kit protocols were adapted for high throughput Tecan liquid handling. Libraries were run on an Illumina NextSeq2000 with paired-end sequencing (2×50 bp). Sample sequencing depth was 8.7 million reads (BRG1), 4.1 million reads (H3K4me3), and 2.4 million reads (IgG) Data were aligned to the hg19 genome using Bowtie2. Data were filtered to remove duplicates, multi-aligned reads, and ENCODE DAC Exclusion List regions.
Figure 1: BRG1 peaks in CUT&RUN.
Figure 2: BRG1 CUT&RUN peak enrichment and functional overlap.
CUT&RUN was performed as described above. BRG1 peaks overlapped with BRD4 antibody CUT&RUN peaks (EpiCypher 13-2003, top), as has been demonstrated in the literature [1]. Gene browser shots were generated using the Integrative Genomics Viewer (IGV, Broad Institute). Three representative loci show overlap of BRG1 peaks with H3K4me3 and BRD4 peaks (bottom).
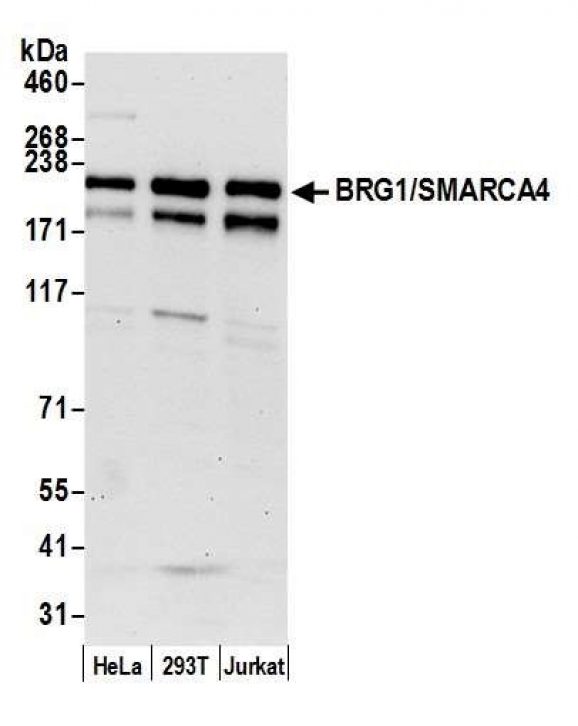
Figure 4: Western blot data.
Figure 6: Immunoprecipitation data.
- Type: Polyclonal
- Host: Rabbit
- Applications: CUT&RUN, IHC, IP, WB
- Reactivity: Human, Mouse
- Format: Antigen affinity-purified
- Target Size: 185 kDa
Immunogen
Between amino acids 75 and 125
Storage
Stable for 1 year at 4°C from date of receipt
Formulation
Antigen affinity-purified antibody in Tris-buffered saline, 0.1% BSA, 0.09% sodium azide
- CUT&RUN: 0.5 µg per reaction
- Immunoprecipitation (IP): 2 – 5 µg/mg lysate
- Immunohistochemistry (IHC): 1:250 – 1:2,000. Epitope retrieval with citrate buffer pH 6.0 is recommended for FFPE tissue sections
- Western Blot (WB): 1:2,000 – 1:10,000
- UniProt ID: P51532
- Gene Name: SMARCA4
- Protein Name: Transcription activator BRG1
- Target Size: 185 kDa
- Alternate Names: BAF190A, BRG1, SNF2B, SNF2L4, ATP-dependent helicase SMARCA4, BRG1-associated factor 190A (BAF190A), Mitotic growth and transcription activator, Protein BRG-1, Protein brahma homolog 1