CUTANA™ Multiomic CUT&RUN Controls Set
In stock
The CUTANA™ Multiomic CUT&RUN Controls Set is a specialized set of controls designed to be paired with the CUTANA™ ChIC/CUT&RUN Kit (EpiCypher 14-1048) to enable direct, simultaneous analysis of DNA methylation and chromatin proteins in a single workflow.
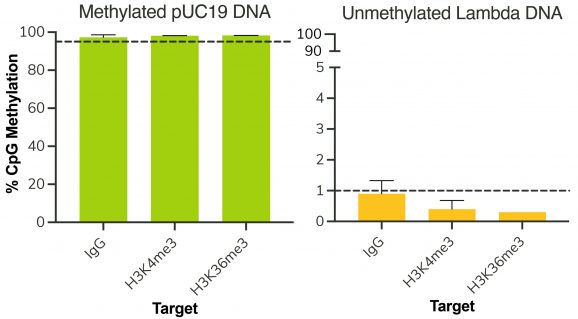
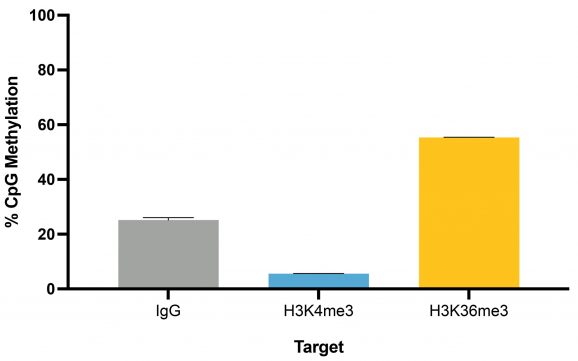
Following isolation of chromatin protein-bound DNA in CUT&RUN, DNA fragments are prepared for sequencing with a cytosine conversion strategy such as Enzymatic Methyl-seq (NEB® EM-seq™, preferred) or bisulfite sequencing to provide base-pair resolution of 5-methylcytosine (5mC). This set includes the essential controls for CUT&RUN followed by EM-seq (CUT&RUN-EM). H3K36me3 antibody is a positive control that enriches DNA associated with active gene bodies, which contain high levels of DNA methylation. H3K36me3 antibody complements the antibodies included in the CUT&RUN kit (H3K4me3, which enriches unmethylated promoters, and IgG, which serves as a negative control) to validate the technical success of the experimental workflow. Pre-fragmented methylated pUC19 and unmethylated Lambda DNAs serve as controls in EM-seq to assess conversion efficiency of unmethylated cytosines.
This controls set is a tailored solution to unlock the capacity of CUT&RUN to generate multiomic insights and investigate crosstalk between DNA methylation and chromatin proteins.
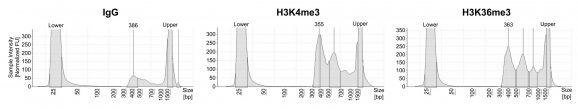
Figure 1: Representative CUT&RUN-EM DNA fragment size distribution analysis
CUT&RUN-EM was performed as described in Figure 6. Library DNA was analyzed by Agilent TapeStation®. This analysis confirmed that mononucleosomes were predominantly enriched in CUT&RUN-EM (~300 bp peaks represent 150 bp nucleosomes+ sequencing adapters). High molecular weight nucleosome laddering is expected with EM-seq and does not affect sequencing quality.
Figure 3: Representative EM-seq conversion efficiency
Figure 4: Representative percent methylation by target
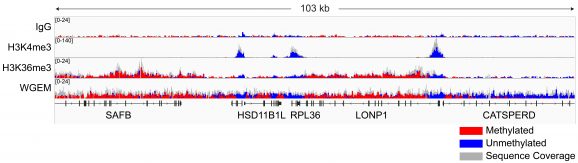
Figure 5: Representative gene browser tracks
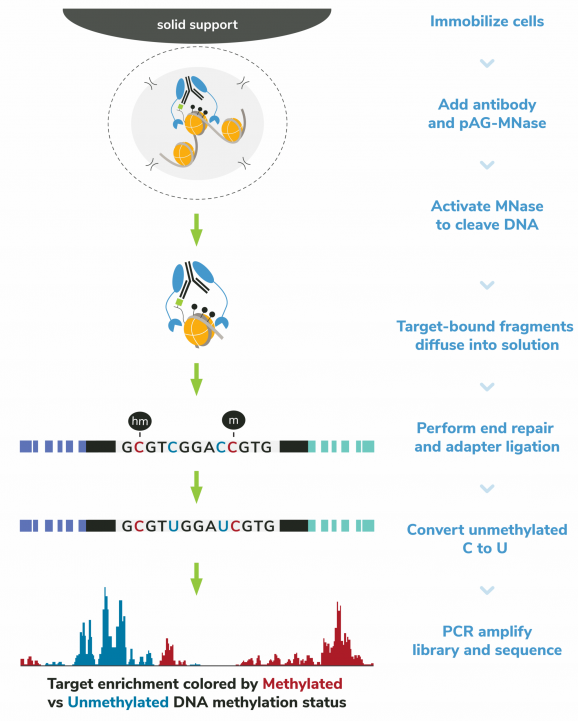
Figure 6: CUT&RUN-EM methods
CUT&RUN-EM was performed using the CUTANA™ Multiomic CUT&RUN Controls Set and the CUTANA™ ChIC/CUT&RUN Kit (EpiCypher 14-1048) starting with 500k K562 cells and 0.5 µg of IgG (EpiCypher 13-0042), H3K4me3 (EpiCypher 13-0060), or H3K36me3 (EpiCypher 13-0058) antibodies. Library preparation was performed with 1 ng of DNA using the NEBNext® Enzymatic Methyl-seq v2 Kit (NEB E8015). Libraries were run on an Illumina NextSeq2000 with paired-end sequencing (2×50 bp). Sample sequencing depth was 31.5 million reads (IgG), 21.3 million reads (H3K4me3), and 20.6 million reads (H3K36me3). Data were aligned to the hg38 genome using Bowtie2. Data were filtered to remove duplicates, multi-aligned reads, and ENCODE DAC Exclusion List regions. Validation data are representative where noted. Refer to the product page for 13-0058 for the lot-specific validation approach for H3K36me3 antibody.
| Item | Cat. No. | QTY |
|---|---|---|
| CUTANA™ H3K36me3 Positive Control Antibody | 13-0058-03 | 8 rxn |
| CUTANA™ CpG Methylated pUC19 Fragmented Control DNA | 18-8001-05 | 20 µL |
| CUTANA™ CpG Unmethylated Lambda Fragmented Control DNA | 18-8002-05 | 20 µL |
| CUTANA™ 0.1X TE Buffer | 21-1025-05 | 2 x 2 mL |
| Item | Cat. No. |
|---|---|
| CUTANA™ ChIC/CUT&RUN Kit | 14-1048/14-1048-24 |
| NEBNext® Enzymatic Methyl-seq v2 Kit | NEB E8015 |