Learn More About EpiCypher’s Custom Nucleosome Assembly Services!
- Ellen Weinzapfel

Chromatin is one of the most rapidly growing fields of biomedical research, with potential applications in diagnostics, prognostics, and therapeutics. The base unit of chromatin structure is the nucleosome, composed of ~147bp DNA wrapped around a histone octamer1. Nucleosomes are the preferred, physiological substrate of many chromatin interacting proteins, and are required to study numerous high-value therapeutic targets, including histone modifying enzymes2-6, chromatin readers7-9 and remodeling complexes10, 11.
EpiCypher is an expert in recombinant and/or semi-synthetic nucleosome assembly, with a diverse collection of recombinant nucleosomes ready-to-order. However, if your nucleosome is not available in our catalog, or if you require a special panel of nucleosomes for your experiments, EpiCypher also offers custom nucleosome assembly services.
EpiCypher Nucleosome Assembly Technologies
As part of these services you will have access to our vast collection of modified histones and DNA templates, as well as support in the development of novel PTMs or DNAs for your project. These technologies are outlined in Figure 1, and include
• EpiDyne® chromatin remodeling substrates : These nucleosomes are manufactured using a longer DNA template, and are modified to include a fluorescent tag (i.e. FRET) or restriction enzyme site, which enables direct monitoring of nucleosome remodeling activity.
• Modified designer nucleosomes, or dNucs™ : dNucs are highly pure, semi-synthetic recombinant nucleosomes, carrying defined histone PTMs that have been confirmed by high-resolution mass spectrometry and western blot.
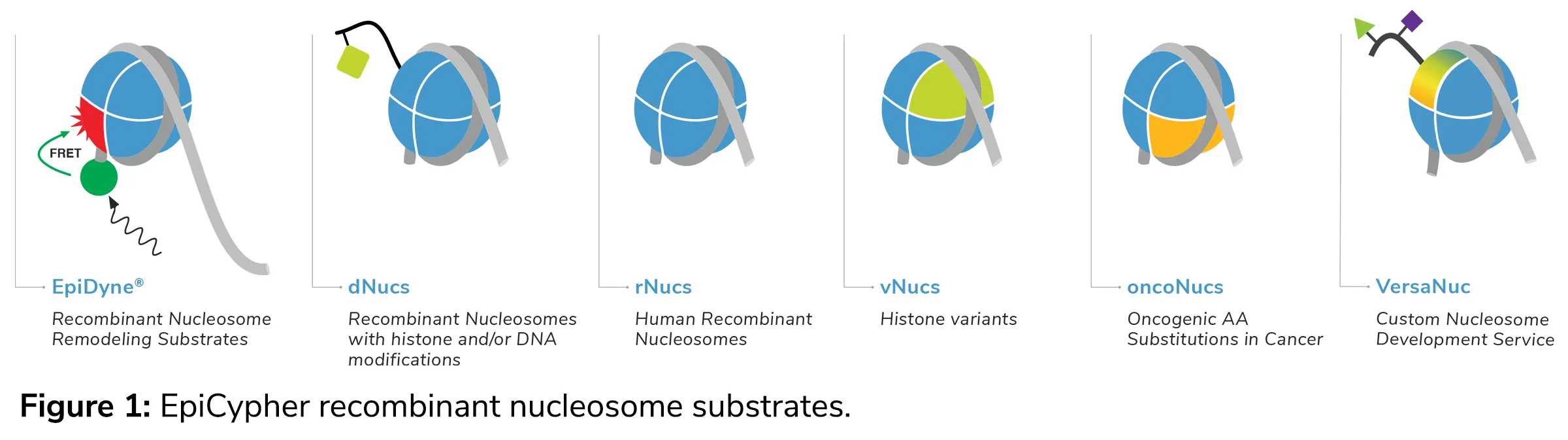
• Recombinant nucleosomes, or rNucs : rNucs are our line of unmodified recombinant nucleosomes, which can be wrapped with a variety of DNA templates for various downstream assays.
• Variant nucleosomes, or vNucs : vNucs are developed using histone octamers that contain unique histone variants, such as H2AZ. Our collection of histone variants can also be incorporated with other technologies for a custom nucleosome assembly (see below).
• Oncogenic nucleosomes, or oncoNucs : oncoNucs are a distinct line of nucleosomes, which carry defined histone amino acid substitution(s) that have been documented in human cancer.
versaNuc™ : Versatile Nucleosome Assembly
One of our most exciting technologies was specifically designed as a personalized nucleosome development platform. The EpiCypher versaNuc™ strategy starts with the assembly of a histone octamer containing a “tailless” H3 histone (truncation of the N-terminal tail at amino acid 32). As outlined in Figure 2, from this starting point, versaNuc provides a wealth of options for customization.
First, you select your DNA, with options ranging from the basic Widom 601 nucleosome positioning sequence12 to longer templates, with the option of adding hemi- or fully-methylated sequences. EpiCypher has many options readily available, and can also work with you to develop specific templates required for your project.
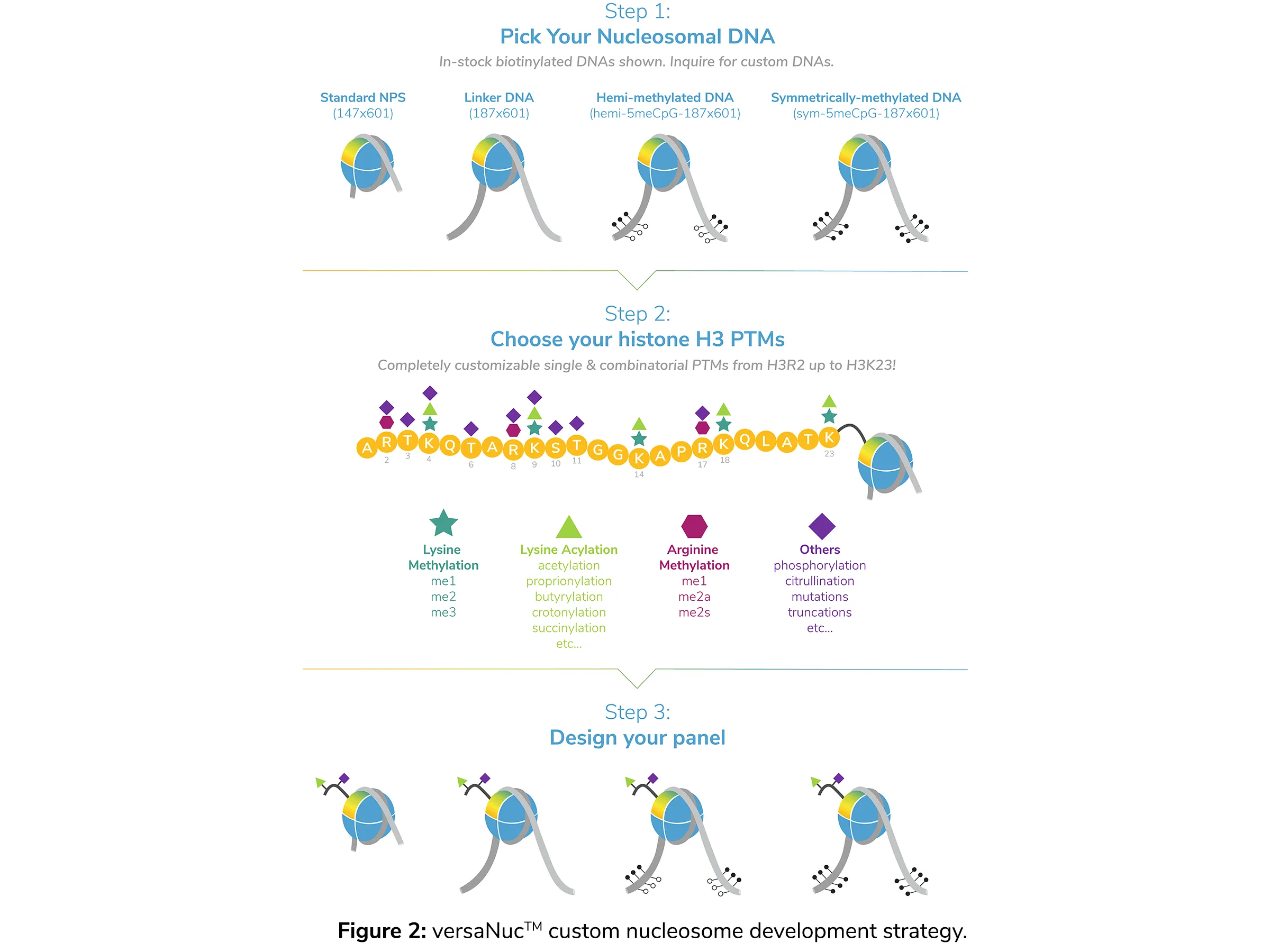
Once the octamer is assembled onto the DNA, you then select the H3 modification(s) you want to add to your nucleosome (Figure 2, middle panel). We can synthesize H3 peptides (amino acids 1-23) carrying single and combinatorial PTMs, including lysine methylation and acylation, arginine methylation, phosphorylation, citrullination, and many more.
These H3 peptides can be directly ligated to the assembled nucleosome, making it feasible to generate entire panels of nucleosomes (or versaNucs) with diverse histone PTMs!
For instance, you could design a core nucleosome with a defined DNA template and “tailless” H3. This nucleosome could be ligated to a variety of modified H3 peptides, thus generating an entire panel of versaNuc substrates.
Conversely, you could also create a series of nucleosomes with different DNA templates, and ligate to a curated set of H3 peptides, for a more complex versaNuc library. There are countless combinations available!
Importantly, versaNuc can also be integrated with our other nucleosome assembly technologies to incorporate modifications or variants of H2A, H2B, or H4. Thus, the development of versaNuc represents a truly remarkable advancement in nucleosome manufacturing, providing unprecedented access to complex chromatin substrates for next-generation epigenetics research.
Working on a new project and interested in learning more about EpiCypher nucleosome services? Let us know how we can help!
References
1. Margueron R, Reinberg D. Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet, 2010. 11(4): p. 285-96. (PubMed PMID: 20300089) (PMC3760772)
2. Li Y, et al. The target of the NSD family of histone lysine methyltransferases depends on the nature of the substrate. J Biol Chem, 2009. 284(49): p. 34283-95. (PubMed PMID: 19808676) (PMC2797197)
3. Strelow JM, et al. The Use of Nucleosome Substrates Improves Binding of SAM Analogs to SETD8. J Biomol Screen, 2016. 21(8): p. 786-94.(PubMed PMID: 27369108)
4. Gil R, et al. SIRT6 exhibits nucleosome-dependent deacetylase activity. Nucleic Acids Res, 2013. 41(18): p. 8537-45.(PubMed PMID: 23892288) (PMC3794599)
5. Yang M, et al. Structural basis for CoREST-dependent demethylation of nucleosomes by the human LSD1 histone demethylase. Mol Cell, 2006. 23(3): p. 377-87.(PubMed PMID: 16885027) (PMC3794599)
6. McGinty RK, et al. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature, 2008. 453(7196): p. 812-6. (PubMed PMID: 18449190) (PMC3774535)
7. Weinberg DN, et al. The histone mark H3K36me2 recruits DNMT3A and shapes the intergenic DNA methylation landscape. Nature, 2019. 573(7773): p. 281-6. (PubMed PMID: 31485078) (PMC6742567)
8. Morrison EA, et al. The conformation of the histone H3 tail inhibits association of the BPTF PHD finger with the nucleosome. Elife, 2018. 7: p. (PubMed PMID: 29648537)(PMC5953545)
9. Nguyen UT, et al. Accelerated chromatin biochemistry using DNA-barcoded nucleosome libraries. Nat Methods, 2014. 11(8): p. 834-40. (PubMed PMID: 24997861) (PMC4130351)
10. Dann GP, et al. ISWI chromatin remodellers sense nucleosome modifications to determine substrate preference. Nature, 2017. 548(7669): p. 607-11. (PubMed PMID: 28767641) (PMC5777669)
11. Lee HS, et al. A cooperative activation loop among SWI/SNF, gamma-H2AX and H3 acetylation for DNA double-strand break repair. EMBO J, 2010. 29(8): p. 1434-45. (PubMed PMID: 20224553) (PMC2868568)
12. Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol, 1998. 276(1): p. 19-42. (PubMed PMID: 9514715)
