Finding the Best Substrate for Studying Histone Modifications
- Ellen Weinzapfel

The Function of Histone Modifications : A Histone Code
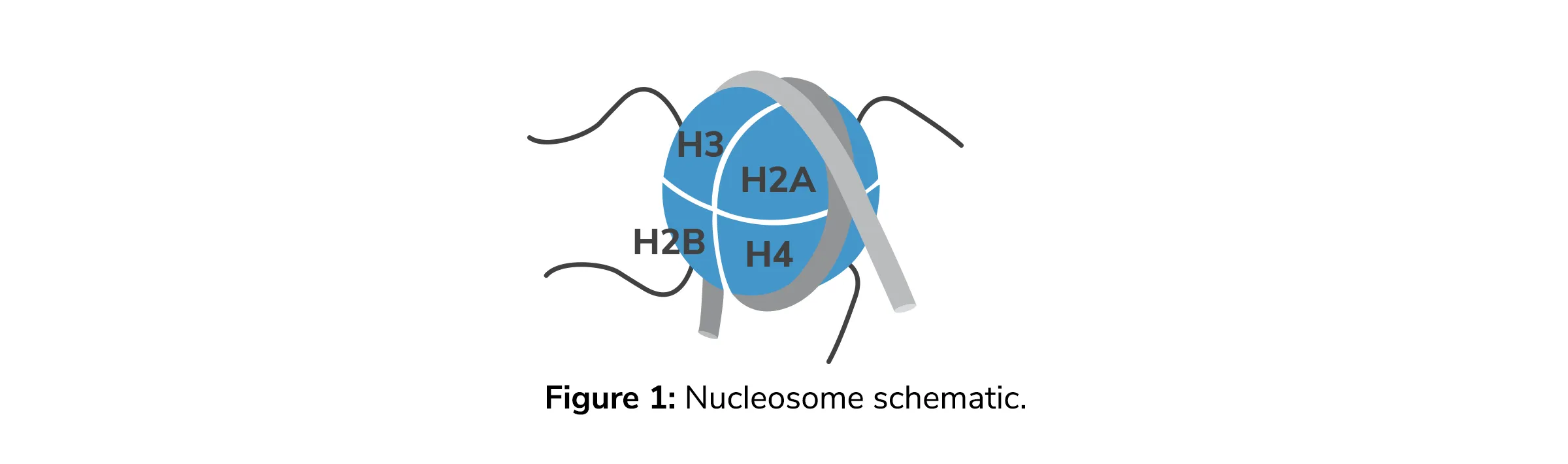
The Ideal Substrate for the Study of Histone Modifications
Mounting evidence suggests that nucleosomes are the optimal substrates to characterize many chromatin regulators in vitro.This is in contrast to previous work, which often used modified histone peptides to identify chromatin inhibitors or define novel histone PTM interactions.
We have covered this topic in a previous blog post regarding the use of histone peptides vs. nucleosomes for histone PTM antibody validation. However, these issues are also relevant for studies of chromatin modifying enzymes. For example, the NSD2 methyltransferase (associated with oncogenic reprogramming in multiple myeloma) requires nucleosomal substrates for in vitro activity 8. Similarly, the SETD8 methyltransferase displays a 10,000-fold decrease in Km for the SAM cofactor when using recombinant nucleosomes vs. histone peptide substrates 9.
The advent of recombinant nucleosomes carrying fully defined histone modifications enables novel or dramatically improved approaches (e.g. reader binding, enzymatic assays, antibody profiling) for next-generation chromatin research.
How are Recombinant Nucleosomes Made?
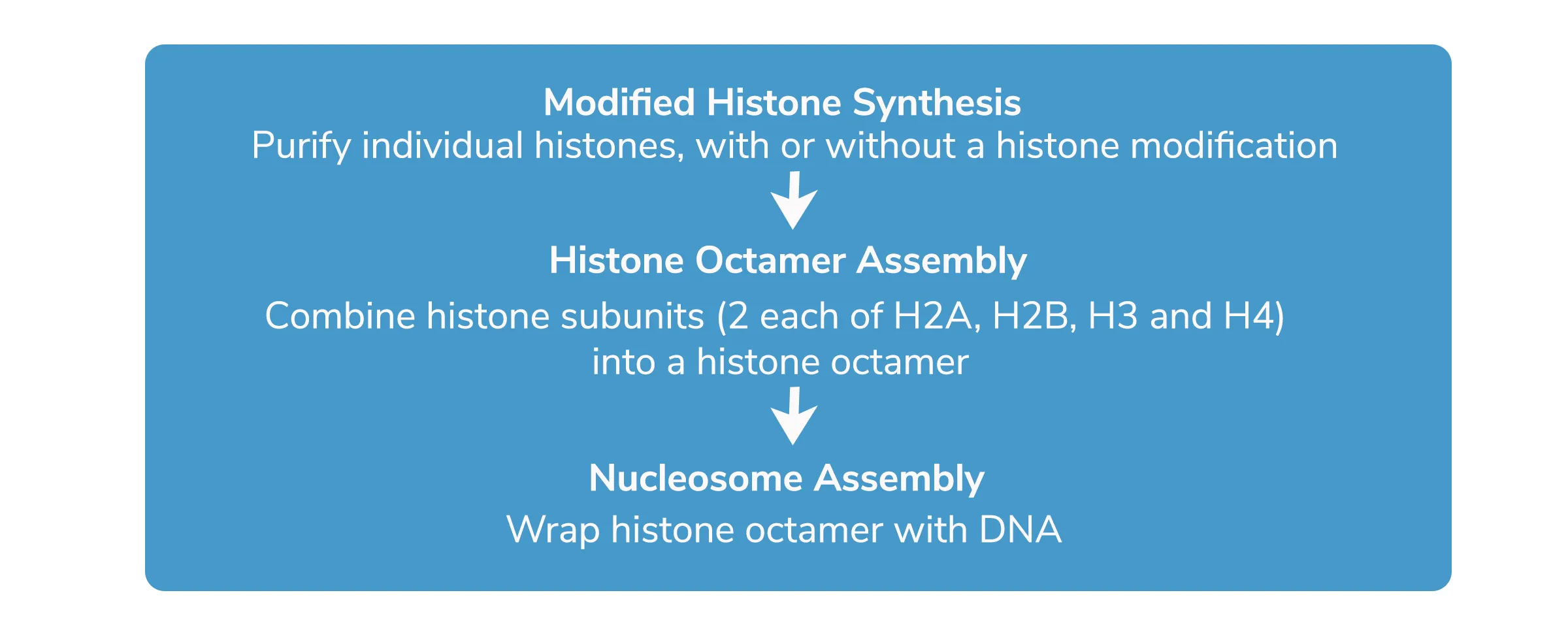
How To Judge a Recombinant Nucleosome
If you are using a recombinant nucleosome for your chromatin experiments, here are a few points to consider:
1) The approach used to develop the modified histones. It is important to ensure that the desired PTMs are incorporated using scar-less methodologies that recapitulate the native histone structure. EpiCypher generates modified histones using several different methods, all of which result in scar-less incorporation of histone modifications.
Why is this important? Many commercially available recombinant nucleosomes are constructed using histone PTM analogs, such as methyl lysine analogs (MLAs) 10, which result in changes to the amino acid sequence at the site of the modification. These unnatural histone modifications analogs have been shown to disrupt interactions with chromatin regulating proteins and histone PTM-specific antibodies, and are not ideal for studying physiological mechanisms. Thus, nucleosomes synthesized with these approaches should be used with extreme caution 11-13.
2) The purity of the modified histone. Rigorous quality control of modified histones is the next step in the assembly of fully defined and homogenous nucleosomes.
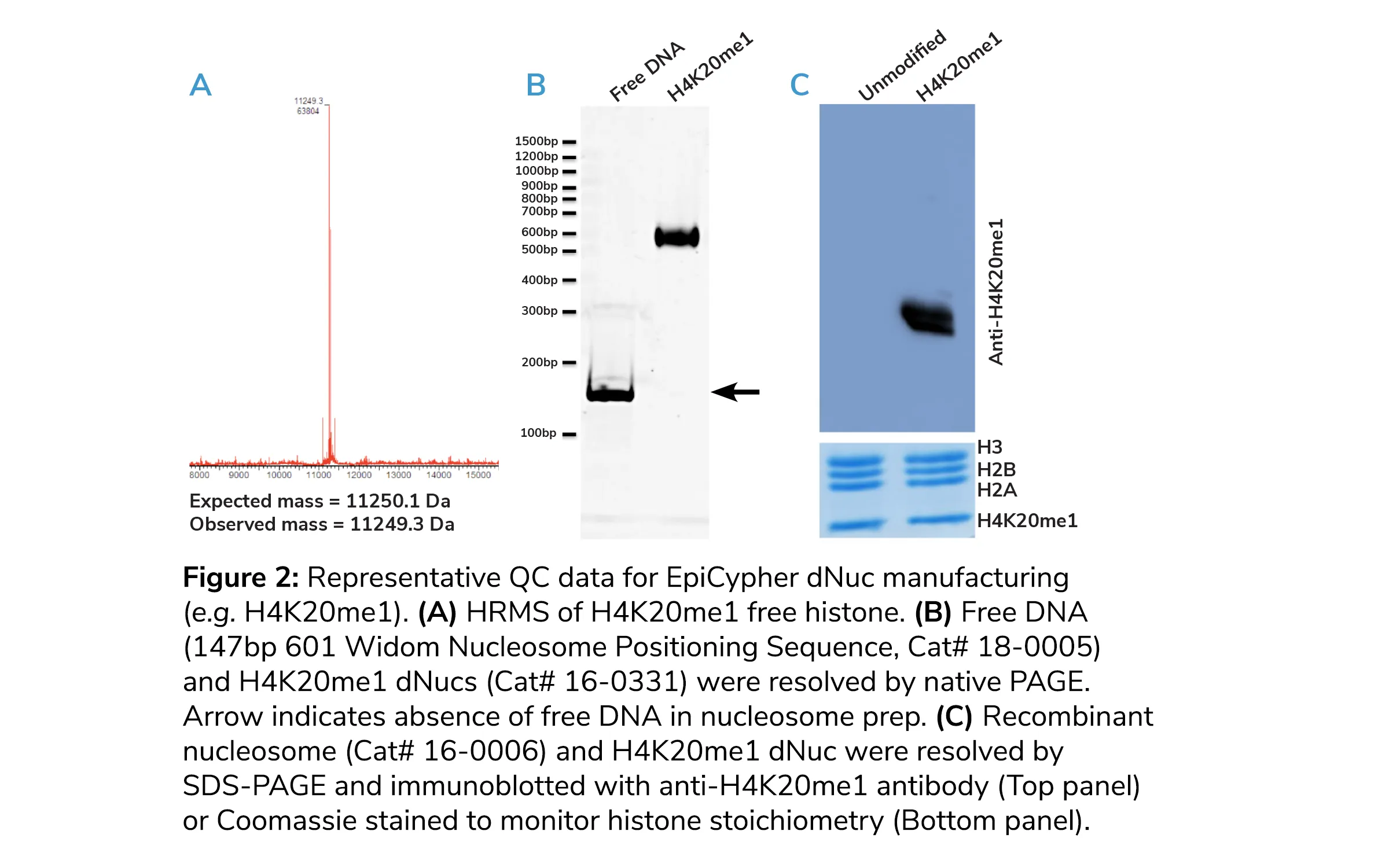
HPLC traces should show a single eluting species, indicating >95% histone purity. Parallel high-resolution mass spectrometry (HRMS) should exhibit a single peak within 1 dalton of expected mass, with no additional mass to charge (m/z) signals of any significance (e.g. Figure 2A). All EpiCypher modified histones are validated by analytical HPLC and HRMS.
Why is this important? Undesirable species (e.g. methionine oxidation) may induce structural alterations, and impact electrostatic or hydrophobic interactions, thus compromising the efficiency of downstream nucleosome assembly.
3) Quality control (QC) metrics of assembled nucleosomes. Quality validation of nucleosomes following DNA assembly is essential to confidence in the final product.
EpiCypher analyzes dNuc assembly on DNA using native PAGE, where an efficient assembly using ~150bp DNA should yield only a single species, with reduced mobility relative to unassembled free DNA (Figure 2B, bottom panel).
Why is this important? Contaminating free DNA can induce an aberrant activity in chromatin modifying enzymes (e.g. NSD2) and must be avoided. Moreover, suboptimal assembly can result in a heterogeneous sample mix, including mispositioned nucleosome species.
We also perform SDS-PAGE of final dNucs to ensure equal stoichiometry of all four histones using Coomassie staining (Figure 2C, bottom panel). We then confirm the presence of the incorporated histone PTM via immunoblot (Figure 2C, top panel).
Why is this important? Contamination with additional protein species or deviation from a 1:1:1:1 ratio could indicate histone degradation or poor assembly, so it is imperative to adequately resolve each histone. Immunoblot is important to determine that the modification is present at the correct location on the histone.
With all parts at play, EpiCypher has an impressive catalog with 83 unique recombinant nucleosomes ready in stock, and also has the capacity to make custom designer nucleosomes with guaranteed high quality and fast turnaround times to help advance your research.
Want to learn more about EpiCypher’s recombinant nucleosome technology and products? Fill out this form or click the button below, and we will be in touch!
References
1. Valencia AM, Kadoch C. Chromatin regulatory mechanisms and therapeutic opportunities in cancer. Nat Cell Biol, 2019. p. (PubMed PMID: 30602726)
2. Luger K, et al. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature, 1997. 389(6648): p. 251-60. (PubMed PMID: 9305837)
3. Jenuwein T, Allis CD. Translating the histone code. Science, 2001. 293(5532): p. 1074-80. (PubMed PMID: 11498575)
4. Rothbart SB, Strahl BD. Interpreting the language of histone and DNA modifications. Biochim Biophys Acta, 2014. 1839(8): p. 627-43. (PubMed PMID: 24631868) (PMC4099259)
5. Cao R, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science, 2002. 298(5595): p. 1039-43. (PubMed PMID: 12351676)
6. Liang G, et al. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc Natl Acad Sci U S A, 2004. 101(19): p. 7357-62. (PubMed PMID: 15123803) (PMC409923)
7. Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell, 2006. 125(2): p. 315-26. (PubMed PMID: 16630819)
8. Li Y, et al. The target of the NSD family of histone lysine methyltransferases depends on the nature of the substrate. J Biol Chem, 2009. 284(49): p. 34283-95.(PubMed PMID: 19808676)(PMC2797197)
9. Strelow JM, et al. The Use of Nucleosome Substrates Improves Binding of SAM Analogs to SETD8. J Biomol Screen, 2016. 21(8): p. 786-94.(PubMed PMID: 27369108)
10. Simon MD, et al. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell, 2007. 128(5): p. 1003-12. (PubMed PMID: 17350582)(PMC2932701)
11. Gloss LM, Kirsch JF. Decreasing the basicity of the active site base, Lys-258, of Escherichia coli aspartate aminotransferase by replacement with gamma-thialysine. Biochemistry, 1995. 34(12): p. 3990-8. (PubMed PMID: 7696264)
12. Gellman SH. On the role of methionine residues in the sequence-independent recognition of nonpolar protein surfaces. Biochemistry, 1991. 30(27): p. 6633-6. (PubMed PMID: 2065050)
13. Seeliger D, et al. Quantitative assessment of protein interaction with methyl-lysine analogues by hybrid computational and experimental approaches. ACS Chem Biol, 2012. 7(1): p. 150-4. (PubMed PMID: 21991995) (PMC3265130)
