Rethink DNA Methylation Profiling with CUTANA™ meCUT&RUN
- Aaron Alcala, PhD

DNA methylation is a cornerstone of epigenetic regulation — and mapping it accurately across the genome is essential for understanding development, disease, and drug mechanisms. But balancing comprehensive, genome-wide DNA methylation profiling with affordability and input requirements continues to be a challenge.
Whole-genome DNA methylation profiling assays, like bisulfite sequencing, demand massive sequencing depths and high costs, while lower-cost alternatives, like DNA methylation microarrays or reduced representation bisulfite sequencing (RRBS), only detect a small fraction of 5-methylcytosine (5mC). For many researchers, those costs and constraints are unsustainable.
That is why EpiCypher developed CUTANA™ meCUT&RUN: A next-generation DNA methylation profiling assay that delivers whole-genome coverage with low input, low cost, and high confidence.
A Cost Effective, Low-Input Method to Map DNA Methylation
CUTANA™ meCUT&RUN delivers high-quality 5-methylcytosine profiles using as few as 10,000 K562 cells and just 15 million reads per sample—without bisulfite conversion (Figure 1). Whether you’re working with clinical biopsies or in vitro models, meCUT&RUN delivers robust and reproducible results with minimal material.

Figure 1: CUTANA meCUT&RUN cell titration experiment using K562 cells.
A Streamlined Workflow with Built-in Flexibility
At the heart of meCUT&RUN is a simple and powerful protocol:
- Use a GST-tagged MeCP2 protein to enrich methylated DNA
- Leverage the CUT&RUN platform for precise, low-background profiling
- Choose your output:
- Standard DNA library prep for ~150 bp resolution (~15–20M reads)
- Or add enzymatic methylation conversion for base-resolution 5mC mapping (~30–50M reads)
Either way, you’ll avoid bisulfite treatment, reduce DNA damage, and simplify your workflow.

Figure 2: meCUT&RUN offers a simple, scalable workflow for DNA methylation sequencing
Save on Sequencing—Not on Coverage
How does meCUT&RUN measure up to whole-genome sequencing strategies? We compared CUTANA meCUT&RUN (standard library prep) and whole-genome DNA methylation sequencing (EM-seq™) across a 97 kb region. meCUT&RUN detects the majority of methylated CpGs with strong signal correlation to EM-seq and significantly reduced sequencing requirements (Figure 3). That’s powerful performance, without the price tag.

Figure 3: meCUT&RUN and EM-seq representative tracks from 500,000 K562 cells. Enzymatic Methyl Seq (EM-seq™) is a trademarked term by New England Biolabs®.
To further assess 5-methylcytosine coverage, we paired meCUT&RUN with enzymatic methylation conversion (meCUT&RUN-EM), enabling direct comparisons to whole-genome DNA methylation sequencing assays. CUTANA™ meCUT&RUN captures approximately 80% of the unique CpGs detected by whole-genome DNA methylation sequencing (EM-seq) using just 20–30 million reads (Figure 4). This high-efficiency performance enables genome-wide methylation profiling from low input samples while reducing sequencing costs and data analysis burden.
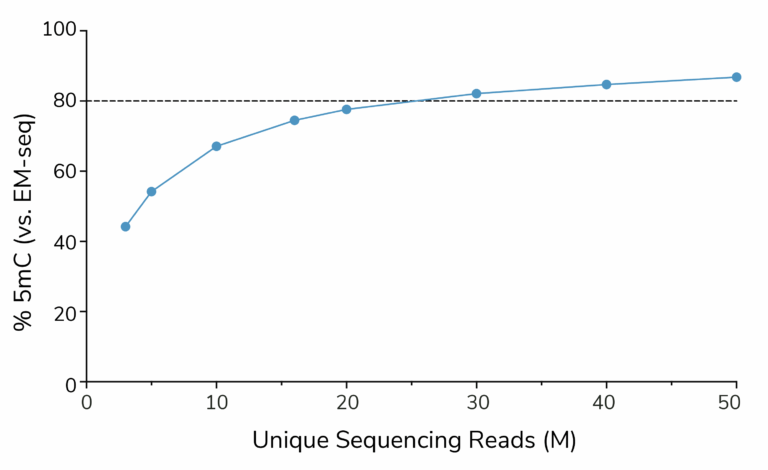
Figure 4: meCUT&RUN was paired with EM-seq for base-pair resolution 5mC analysis (meCUT&RUN-EM). Data were downsampled to 3 to 50 M uniquely aligned reads. The number of methylated CpG positions was calculated for each downsampled dataset and normalized to the number of 5mC position detected by EM-seq.
Improved 5-Methylcytosine Detection vs. RRBS
We also compared meCUT&RUN to Reduced Representation Bisulfite Sequencing (RRBS), a targeted DNA methylation sequencing strategy. While RRBS provides sparse, biased coverage due to its enzymatic digestion and size-selection steps, meCUT&RUN-EM delivers genome-wide signal with low sequencing requirements (Figure 5A).
In addition, meCUT&RUN captures 5mCs across a broad range of functional elements (Figure 5B):
- 100% of 5mC at TSS and CpG islands
- Over 70% of 5mCs at enhancers, gene bodies, and repeat elements
The balanced coverage of meCUT&RUN provides a more complete view of the methylome vs. RRBS, enabling deeper insight into gene regulation, epigenetic memory, and disease-relevant loci.

Figure 5: CUTANA meCUT&RUN was paired with enzymatic conversion to thoroughly evaluate its performance against targeted bisulfite-conversion assays (RRBS) and whole-genome, enzymatic-based approaches (EM-seq; 300 M reads) in K562 cells. (A) Representative sequencing tracks. (B) Percent 5mC coverage calculated relative to EM-seq.
Get Started with meCUT&RUN
The CUTANA™ meCUT&RUN Kit contains everything you need to enrich and profile methylated DNA. We also offer standalone reagents for flexible experimental design:
- GST-MeCP2: Binds methylated DNA
- Anti-GST Tag Antibody: Enables targeted cleavage in meCUT&RUN
- Fragmented DNA Controls: Spike-ins for base-pair resolution 5mC profiling
