SNAP-CUTANA™ HA Tag Panel
In stock
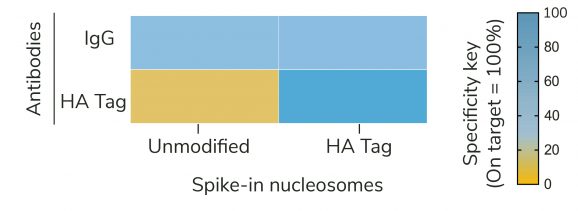
The SNAP-CUTANA™ HA Tag Panel of spike-in controls for CUT&RUN offers an in-assay control to validate anti-HA antibodies and confirm the success of CUT&RUN reactions involving HA epitope-tagged chromatin proteins. This essential positive control guides troubleshooting to differentiate problems with HA epitope-tagging (including transgene expression, chromatin binding of the tagged protein, solvent accessibility of the tag, etc.) from technical failures in the CUT&RUN workflow. The panel consists of two nucleosomes containing unmodified histone H3 or 3xHA-H3 fusion, each wrapped with two uniquely barcoded DNA templates (A and B, for an internal technical replicate). The nucleosomes are individually conjugated to paramagnetic beads and pooled into a single panel for convenient one-step spike-in to CUT&RUN reactions. The panel is added alongside ConA-immobilized cells just prior to the addition of anti-HA or IgG negative control antibodies (see Application Notes and Table 1). The release of genomic chromatin and the barcoded nucleosomes by pAG-MNase is dependent on the specificity of the antibody used. After sequencing, the relative read count of recovered HA vs. unmodified nucleosomes provides a quantitative metric of on- vs. off-target recovery (Figure 2), thereby gauging experimental success and guiding troubleshooting efforts. See the most recent CUTANA™ CUT&RUN protocol and SNAP-CUTANA™ Spike-in User Guide for detailed information on workflow integration, expected results, data analysis, and troubleshooting.
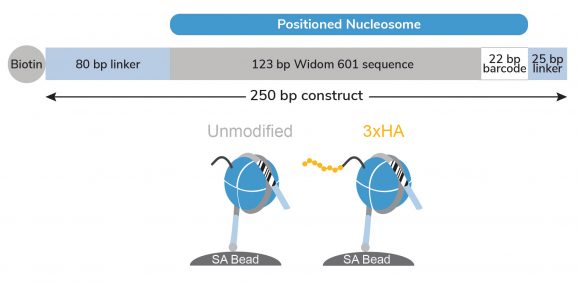
Figure 1: Schematic of SNAP-CUTANA™ HA Tag Panel
The HA Tag Panel contains two nucleosomes – one has an H3 tail fusion to a 3xHA Tag epitope and one is an unmodified control. Both octamers are wrapped with two uniquely barcoded DNA templates (A and B). Each 250 bp DNA template contains a 123 bp 601 nucleosome positioning sequence (gray) [1], a unique 22 bp DNA-barcode (white; 4 barcodes total), and a 5’ biotin-TEG. The 5’ and 3’ linkers (blue) are compatible with cleavage by pAG-MNase (EpiCypher 14-1048, 15-1016) during CUT&RUN. The nucleosomes are individually pre-conjugated to paramagnetic beads and pooled for convenient use.
Figure 2: SNAP-CUTANA™ HA Tag Panel provides an in-assay control for CUT&RUN reactions targeting HA-tagged proteins
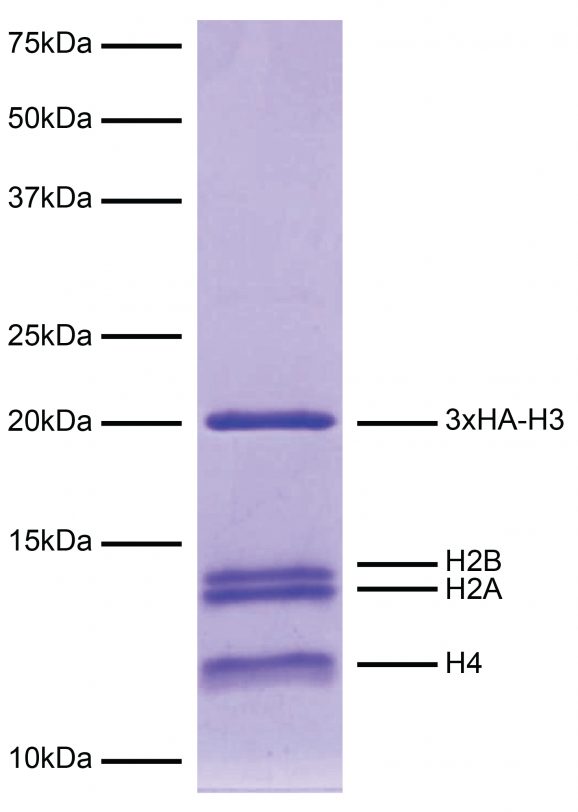
Figure 4: Protein gel data
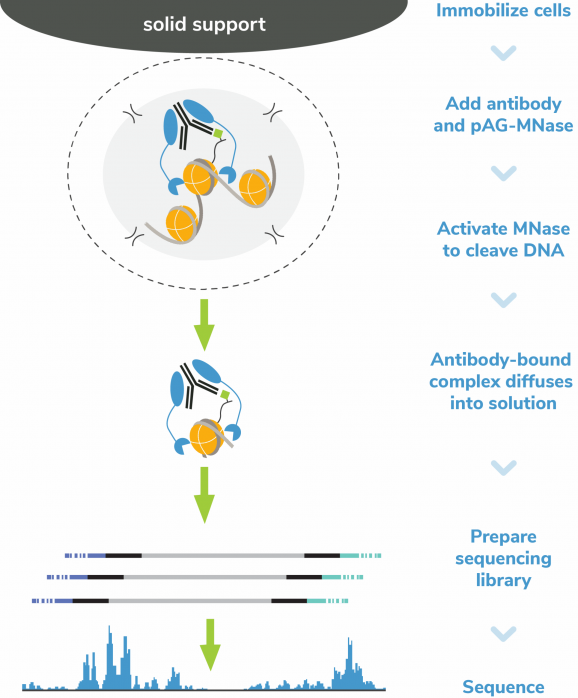
Figure 5: CUT&RUN methods
CUT&RUN was performed on 500k MDA-MB-231 native cells stably expressing 3xHA-tagged GATA3 [1]* using the CUTANA™ ChIC/CUT&RUN Kit v3 (EpiCypher 14-1048). SNAP-CUTANA™ HA Tag Panel was added just prior to the addition of either HA Tag (0.5 µg; EpiCypher 13-2010) or IgG negative control (0.5 µg; EpiCypher 13-0042) antibodies. Library preparation was performed with 5 ng of DNA (or the total amount recovered if less than 5 ng) using the CUTANA™ CUT&RUN Library Prep Kit (EpiCypher 14-1001/14-1002). Libraries were run on an Illumina NextSeq2000 with paired-end sequencing (2×50 bp). Data were aligned to the hg19 genome using Bowtie2. Data were filtered to remove duplicates, multi-aligned reads, and ENCODE DAC Exclusion List regions.
*Thanks to Dr. Takaku (UND) for 3xFLAG-GATA3-3xHA MDA-MB-231 cells.
To resuspend beads, gently mix into an even suspension by pipetting; DO NOT VORTEX
Formulation
A mixture of two semi-synthetic nucleosomes conjugated to paramagnetic beads in 10 mM sodium cacodylate pH 7.5, 100 mM NaCl, 1 mM EDTA, 50% glycerol (w/v), 1x Protease Inhibitor Cocktail, 100 μg/mL BSA, 10 mM β-mercaptoethanol.
Application Notes
See the most recent CUTANA™ CUT&RUN protocol and SNAP-CUTANA™ Spike-in User Guide for detailed information on workflow integration, expected results, data analysis, and troubleshooting. In Brief:
Product Use: Use the SNAP-CUTANA™ HA Tag Panel for reactions containing HA and IgG antibodies. Just before antibody addition in CUT&RUN, gently pipette to resuspend beads (do not vortex), then spike in 2 μL per 500k cells. If using less than the standard number of cells, decrease the amount of SNAP-CUTANA spike-in linearly by preparing a “working stock” dilution of the panel in Antibody Buffer, made fresh the day of use (Table 1). Adjust spike-in volume as needed aiming for the spike-in barcodes to comprise ~1% of the total unique sequencing reads. Table 1 gives recommended dilution amounts for varying numbers of starting cells, but optimization may be required for user-specific conditions.
Data Analysis: Detailed instructions are in the SNAP-CUTANA™ Spike-in User Guide (see Documents & Resources section below). Perform paired-end sequencing for a minimum of 50 bases. The Widom 601 DNA and DNA barcodes are distinct from human, mouse, fly, and yeast genomes such that they can be readily distinguished from sample chromatin. A shell script (.sh file extension) for spike-in alignment and an excel template for heatmap generation are available in the Documents & Resources section below. The shell script can be opened with any basic text editor program and contains detailed instructions hashed (#) at the beginning of the document. Make sure to copy and paste the R1 & R2 echo loop so there is a set for each reaction being analyzed.