CUTANA™ Non-Hot Start 2X PCR Master Mix for CUT&Tag
In stock
CUTANA Non-Hot Start 2X PCR Master Mix for CUT&Tag is specifically formulated to amplify next generation sequencing (NGS) libraries from CUTANA™ pAG-Tn5 tagmented DNA generated during CUT&Tag (EpiCypher 15-1017) [Kaya-Okur et al., 2019]. This ready-to-use master mix only requires the addition of primers and the tagmented chromatin to achieve high fidelity amplification of NGS libraries.
NOTE: this product was previously named “CUTANA™ High Fidelity 2X PCR Master Mix”
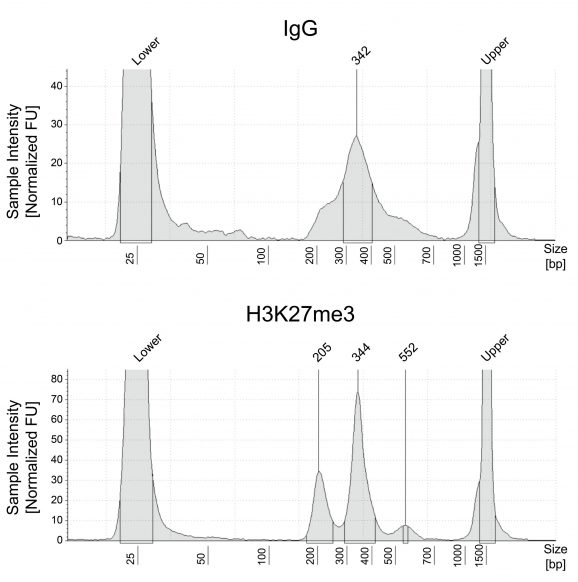
Figure 1: CUT&Tag Next-Generation Sequencing (NGS) libraries
CUT&Tag was performed as described in Figure 3. Recovered DNA was directly PCR amplified to produce sequence-ready libraries. Agilent TapeStation traces for libraries derived from negative control Rabbit IgG (top) and H3K27me3 (bottom) are shown. Excised DNA is highly enriched for mononucleosomes (peak at ~300 bp reflects ~150 bp insert size).
Figure 2: CUT&Tag NGS data
CUT&Tag was performed as described in Figure 3. A 116 kb representative locus is shown for IgG, H3K4me3, or H3K27me3 antibodies. NGS libraries amplified using the CUTANA Non-Hot Start 2X PCR Master Mix represented the expected genomic distribution of H3K4me3 and H3K27me3. Image was generated using the Integrative Genomics Viewer (IGV, Broad Institute).
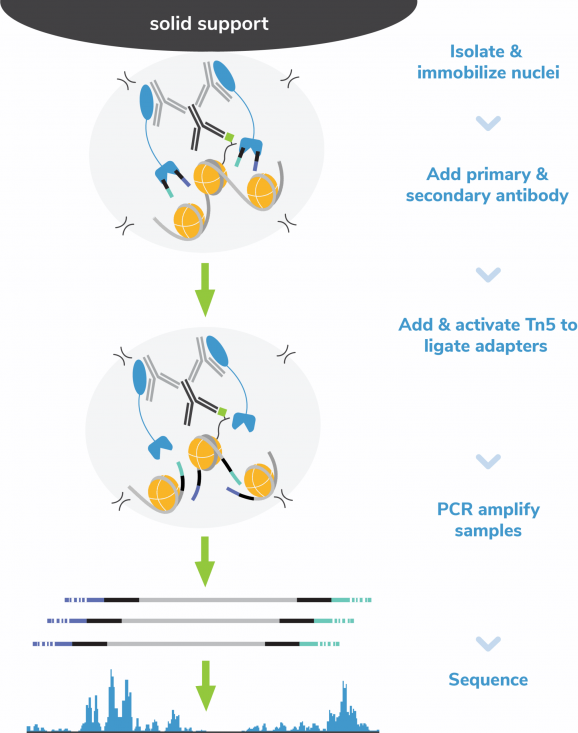
Figure 3: CUT&Tag methods
CUT&Tag was performed on 100k K562 nuclei with 0.5 µg of either IgG (EpiCypher 13-0042), H3K4me3 (EpiCypher 13-0041), or H3K27me3 (ThermoFisher MA5-11198) antibodies following the EpiCypher Direct-to-PCR CUT&Tag protocol. CUT&Tag DNA was directly PCR amplified using CUTANA Non-Hot Start 2X PCR Master Mix to produce NGS libraries. Libraries were run on an Illumina NextSeq2000 with paired-end sequencing (2×50 bp). Sample sequencing depth was 0.3 million reads (IgG), 1.7 million reads (H3K4me3), and 3.8 million reads (H3K27me3). Data were aligned to the hg19 genome using Bowtie2. Data were filtered to remove duplicates, multi-aligned reads, and ENCODE DAC Exclusion List regions.
- Type: Polymerase
- Source: E. coli
- Epitope Tag: None
Application Notes
Recommended use: For detailed instructions regarding non-hot start PCR cycling conditions in CUT&Tag, see CUTANA™ Direct-to-PCR CUT&Tag protocol.
NOTE: Not recommended for use with primers or templates containing uracil.