SNAP-ChIP® K-MetStat™ Panel
A panel of distinctly modified mononucleosomes assembled from recombinant human histones expressed in E. coli (two each of histones H2A, H2B, H3 and H4; accession numbers: H2A-P04908; H2B-O60814; H3.1-P68431 or H3.2-Q71DI3*; H4-P62805) wrapped by 147 base pairs of barcoded Widom 601 positioning sequence DNA. The panel is comprised of a pool of 1 unmodified plus 15 histone H3 or H4 post-translational modifications (PTMs, created by a proprietary semi-synthetic method): H3K4me1, H3K4me2, H3K4me3, H3K9me1, H3K9me2, H3K9me3, H3K27me1, H3K27me2, H3K27me3, H3K36me1, H3K36me2, H3K36me3, H4K20me1, H4K20me2 and H4K20me3. Each distinctly modified nucleosome is distinguishable by a unique sequence of DNA (“barcode”) at the 3′ end that can be deciphered by qPCR or next-generation sequencing. Each of the 16 nucleosomes in the pool is wrapped by 2 distinct DNA species, each containing a distinct barcode (“A” and “B”, see SNAP-ChIP Manual) allowing for an internal technical replicate.
* Histone H3.2 contains a Cys to Ala mutation at position 110.
Background References:
Grzybowski A et al. 2015 Mol Cell 58: 886 – 899 LINK
DNA Gel Data
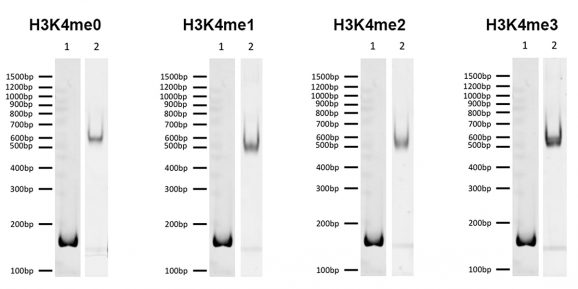
Representative images for SNAP-ChIP K-MetStats (H3K4me0 = unmodified, H3K4me2, H3K4me3) run on a native PAGE gel and stained with ethidium bromide to visualize DNA. Lane 1: Free 147bp DNA used in nuclesome assembly (100 ng). Lane 2: Intact nucleosomes (200 ng) showing lack of free DNA. Identical experiments were performed for the entire K-MetStat Panel.
Protein Gel Data
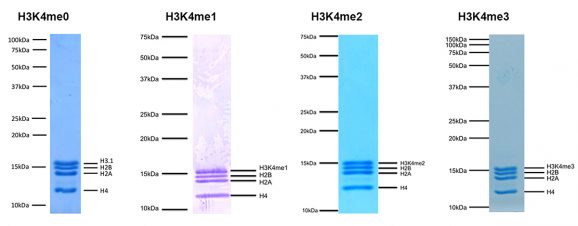
Representative Coomassie stained PAGE gel for SNAP-ChIP K-MetStats (2 µg each of unmodified, H3K4me1, H3K4me2, H3K4me3) to demonstrate the purity of the histones in the preparation. Sizes of molecular weight markers and positions of the core histones (H2A, H2B, H3 and H4) are indicated. Identical experiments were performed for the remainder of the K-MetStat Panel.
ChIP Data
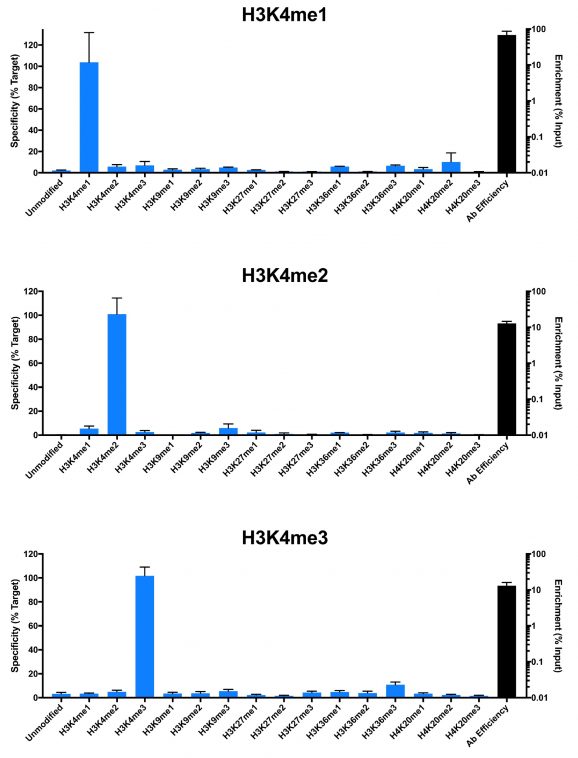
Representative images for SNAP-ChIP K-MetStats (unmodified, H3K4me1, H3K4me2, H3K4me3) assayed in a chromatin immunoprecipitation (ChIP) experiment using commercially available ChIP grade antibodies (3 µg, n = 3). Quantitative Real-Time PCR (qPCR) for the DNA barcodes corresponding to unmodified (H3K4me0), H3K4me1, H3K4me2, and H3K4me3 nucleosomes show recovery of the barcodes corresponding to the expected antibody target. Identical experiments were performed for the remainder of the K-MetStat Panel (H3K9me1, H3K9me2, H3K9me3, H3K27me1, H3K27me2, H3K27me3, H3K36me1, H3K36me2, H3K36me3, H4K20me1, H4K20me2 and H4K20me3).
N.B.-SNAP-ChIP is extremely useful for determining antibody cross-reactivity with off-target modifications. The cross-reactivity observed above (K4me1 & K4me2) is not indicative of a failure in the SNAP-ChIP procedure, but in the antibody to specifically recognize its target.
Purified recombinant mononucleosomes, containing a mixture of 16 (1 unmodified plus 15 unique) H3 and H4 PTMs in 10 mM sodium cacodylate, pH 7.5, 100 mM NaCl, 1 mM EDTA, 50% glycerol (w/v), 1x Protease Inhibitor cocktail, 100 ug/mL BSA, 10 mM β-mercaptoethanol. Molarity = 0.6 nM. MW = ~199382.1 Da (average MW of all 16 nucleosomes).
SNAP-ChIP K-MetStats are highly purified recombinant mononucleosomes and are suitable for use as spike-in controls for ChIP reactions, for antibody specificity testing or for effector protein binding experiments. SNAP-ChIP K-MetStats from EpiCypher do not contain detectable levels of free DNA which could alter assayed activities. See manual for more information. SNAP-ChIP can easily be introduced into existing ChIP and ChIP-seq workflows for experiment normalization and monitoring of technical variability.