dCypher™ Nucleosome Full Panel
1 in stock
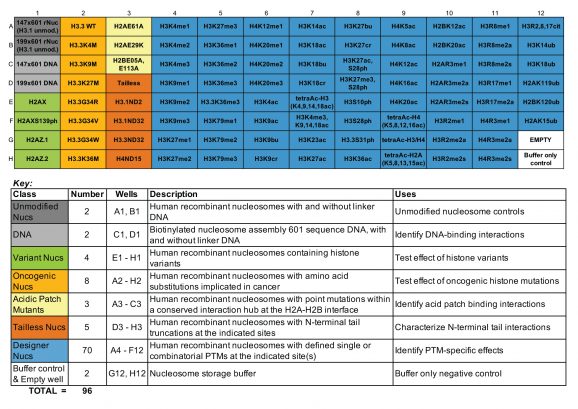
Understanding epigenetic reader proteins and their binding preference for post-translational modifications (PTMs) is key to unveiling how these proteins regulate genome processes and how their dysregulation may be contributing to disease. The dCypher™ Nucleosome Panel enables access to epigenetic diversity in a physiologically relevant context, the nucleosome, where chromatin reader activity can be accurately interrogated. The 96-well plate contains recombinant mononucleosomes as well as appropriate controls. Human histones expressed in E. coli bearing single and combinatorial histone post-translational modifications (PTMs) are wrapped by 147 or 199 base pair DNA with a 5’ biotin-TEG group and a central 601-positioning sequence, identified by Lowary and Widom [1].
All nucleosomes in the panel are subjected to EpiCypher’s rigorous quality control metrics, including: ESI-TOF mass spectrometry analysis of the modified histones, SDS-PAGE to confirm octamer composition and purity, native PAGE to confirm nucleosome assembly, and western blot analysis of the PTM, histone mutation, or histone variant (if applicable). For the full list of nucleosomes in the panel, including individual catalog numbers of full-size (50 µg) products, see the Documents & Resources section for the associated Excel sheet.
Background References:
[1] Lowary & Widom J. Mol. Biol. (1998). PMID: 9514715
[2] Weinberg et al. Nature (2019). PMID: 31485078
[3] Weinberg et al. Nat. Genet. (2021). PMID: 33986537
[4] Dilworth et al. Nat. Chem. Biol (2022). PMID: 34782742
[5] Albanese et al. ACS Chem. Biol. (2020). PMID: 31634430
Product References:
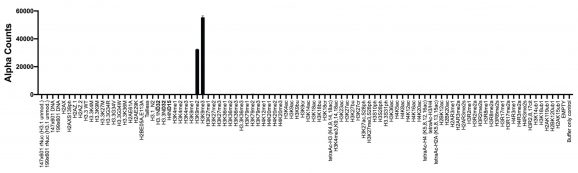
Figure 2: Chromatin reader binding data
GST-tagged HP1β (100 nM, EpiCypher 15-0074) was assayed with AlphaScreen (PerkinElmer) technology to measure binding to nucleosomes (x-axis) in the dCypher nucleosome Panel (10 nM). HP1β shows binding to the H3K9me3 and H3K9me2 dNucs, consistent with its reported binding preference [5].
Gene & Protein Information
- Uniprot ID: H2A – P04908 (alt. names: H2A type 1-B/E, H2A.2, H2A/a, H2A/m)
H2B – O60814 (alt. names: H2B K, HIRA-interacting protein 1)
H3.1 – P68431 (alt. names: H3, H3/a, H3/b, H3/c, H3/d)
H3.2 – Q71DI3
H3.3 – P84243
H4 – P62805
Application Notes
Access to epigenetic diversity in the context of a physiological nucleosome enables broad end-user applications, including nucleosome binding studies (e.g. chromatin reader binding preferences [2-4] see Figure 2), enzyme screening assays (e.g. identification of preferred substrates), and antibody specificity testing (e.g. for applications where histone peptide specificity is an insufficient surrogate). The biotin group on the DNA facilitates a wide variety of applications involving streptavidin capture.