GST-MeCP2 for meCUT&RUN
CUTANA™ GST-MeCP2 for meCUT&RUN is a GST-tagged MeCP2 methyl binding domain that is validated to enrich regions of chromatin with symmetrically methylated CpGs in CUTANA™ meCUT&RUN, a modified CUT&RUN workflow that enables streamlined, high-resolution mapping of DNA methylation. meCUT&RUN can be followed by a traditional library prep method, such as the CUTANA™ CUT&RUN Library Prep Kit (EpiCypher 14-1001/14-1002), to provide ~150 bp resolution profiles of DNA methylation enrichment, or by a cytosine conversion strategy such as Enzymatic Methyl-seq (NEB® EM-seq™, preferred) or bisulfite sequencing to provide base-pair resolution of 5-methylcytosine (5mC).
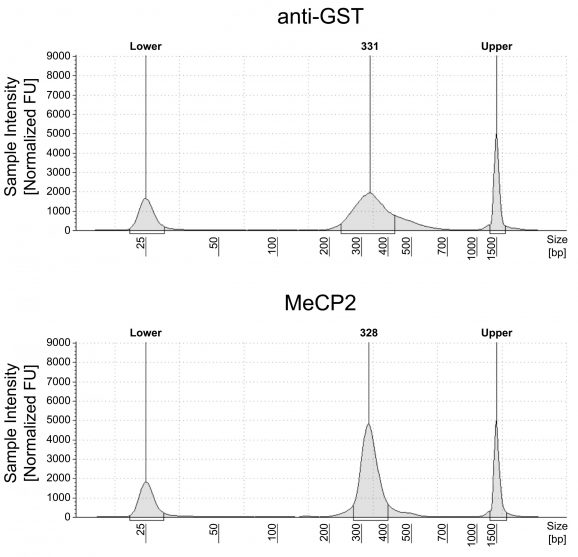
Figure 1: meCUT&RUN DNA fragment size distribution analysis
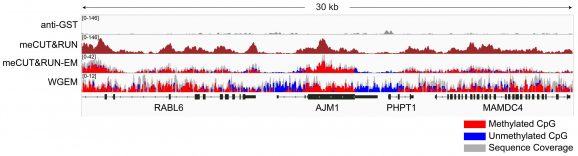
Figure 2: Gene browser tracks
Figure 3: meCUT&RUN methods
meCUT&RUN was performed starting with 500k K562 cells with either 2.5 µL of 20X GST-MeCP2 added as the primary binding reagent or 0.5 µg of a secondary antibody-only control (anti-GST antibody, EpiCypher 13-0073) to determine background cleavage. Library preparation was performed using 5 ng of meCUT&RUN-enriched DNA (or the total amount recovered if less than 5 ng) using the CUTANA™ CUT&RUN Library Prep Kit (EpiCypher 14-1001/14-1002). Libraries were run on an Illumina NextSeq2000 with paired-end sequencing (2×50 bp). Sample sequencing depth was 7.7 million reads (anti-GST) and 8.4 million reads (GST-MeCP2). Data were aligned to the hg38 genome using Bowtie2. Data were filtered to remove duplicates, multi-aligned reads, and ENCODE DAC Exclusion List regions.
| ITEM | CAT. NO. |
|---|---|
| CUTANA™ Anti-GST Tag Antibody | 13-0073 |
| CUTANA™ ChIC/CUT&RUN Kit | 14-1048/14-1048-24 |
| CUTANA™ CUT&RUN Library Prep Kit | 14-1001/14-1002 |
- Prepare Pre-Wash Buffer with 200 mM NaCl*. This is used to prepare Wash Buffer, Digitonin Buffer, and Antibody Buffer. *NOTE: Standard CUT&RUN uses 150 mM NaCl in these buffers; however, excess salt is recommended to improve MeCP2 specificity for methylated DNA.
- In place of a primary antibody, add 2.5 µL of GST-MeCP2 to each reaction, and incubate on a nutator at 4˚C overnight.
- On Day 2, discard the GST-MeCP2 supernatant and resuspend each reaction in 50 µL cold Digitonin Buffer.
- Add 1.0 µL anti-GST Tag Antibody (EpiCypher 13-0073)* to each reaction and incubate on a nutator at room temperature for 30 minutes. *NOTE: Secondary-only (anti-GST Tag) reactions are recommended as a negative control.
- At the end of the incubation, discard the supernatant and wash two times with 200 µL cold Digitonin Buffer.
- Resuspend in 50 µL per reaction cold Digitonin Buffer and proceed to pAG-MNase (EpiCypher 15-1016/15-1116) addition per the standard protocol.
Sequencing Libraries can be prepared using two methods:
Option 1: Traditional library prep, which provides a CUT&RUN-like view of genomic regions with high DNA methylation. The CUTANA CUT&RUN Library Prep Kits (EpiCypher 14-1001/14-1002) are compatible with this approach.
Option 2: A cytosine conversion strategy (e.g., New England Biolabs NEBNext® Enzymatic Methyl-seq v2 Kit, E8015), which provides base-pair resolution of 5mC enrichment. Prior to EM-seq conversion, add CUTANA™ Fragmented Controls for DNA Methylation Sequencing (EpiCypher 14-1804) to each reaction as outlined in the 14-1804 TDS.