HA Tag CUTANA™ CUT&RUN Antibody
In stock
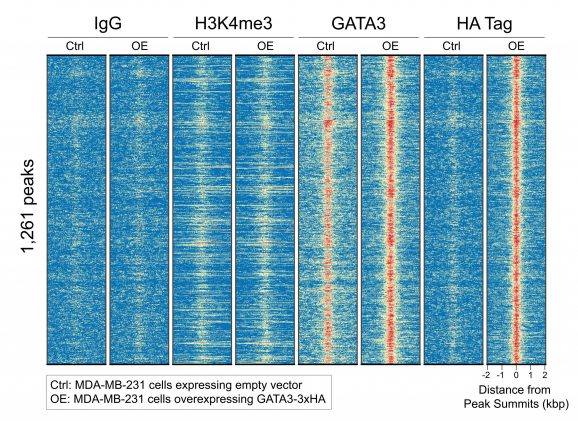
This antibody meets EpiCypher’s “CUTANA Compatible” criteria for performance in Cleavage Under Targets and Release Using Nuclease (CUT&RUN) and/or Cleavage Under Targets and Tagmentation (CUT&Tag) approaches to genomic mapping. Every lot of a CUTANA Compatible antibody is tested in the indicated approach using EpiCypher optimized protocols and determined to yield peaks that show a genomic distribution pattern consistent with reported function(s) of the target. HA antibody is useful for studies utilizing HA-tagged target proteins. HA Tag antibody produces CUT&RUN peaks (Figure 2) in breast cancer cells expressing 3xHA-tagged GATA3 transcription factor [Takaku et al., 2016]*.
Trademarks and copyrights
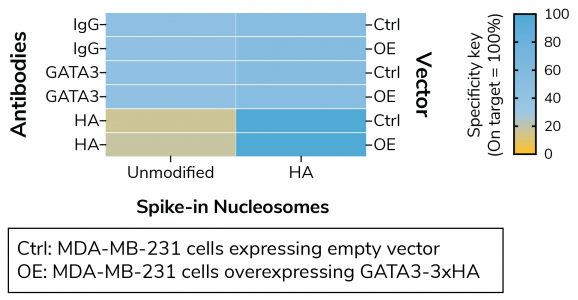
Figure 1: Defined nucleosome spike-ins provide in-assay control for HA Tag antibody in CUT&RUN
CUT&RUN sequencing reads were aligned to the unique DNA barcodes corresponding to each nucleosome in the SNAP-CUTANA™ HA Tag Panel (EpiCypher 19-5002). Data are expressed as a percent relative to on-target recovery (HA Tag set to 100%) or total counts (IgG/GATA3). IgG/GATA3 show no preferential binding to unmodified or HA spike-in nucleosomes. HA Tag antibody selectively enriches the HA Tag spike-in nucleosome, validating the antibody in CUT&RUN.
Figure 2: HA-tagged protein peaks in CUT&RUN
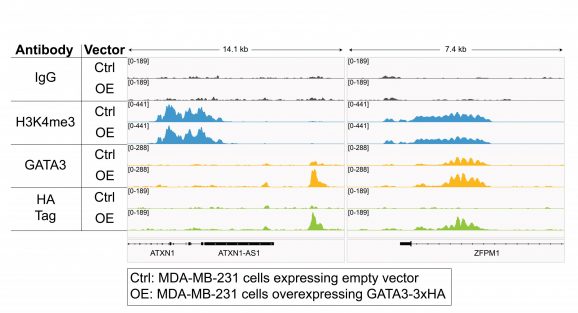
Figure 3: CUT&RUN representative browser tracks for HA-tagged protein
Figure 4: Western blot data
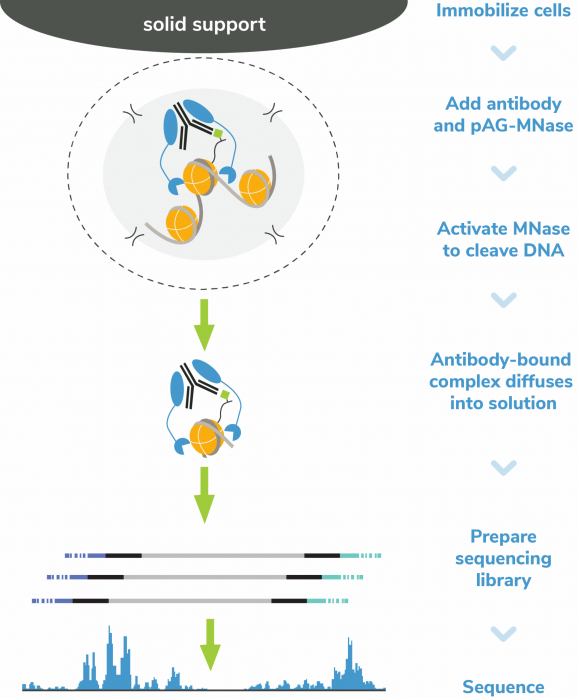
Figure 5: CUT&RUN Methods
CUT&RUN was performed on 500k MDA-MB-231 native cells stably overexpressing c-terminal 3xHA-tagged GATA3 [1] or containing vector control. 0.5 µg of either HA Tag, H3K4me3 (EpiCypher 13-0060), GATA3 (CST 5852), or IgG (EpiCypher 13-0042) antibodies were used with the CUTANA™ ChIC/CUT&RUN Kit v4 (EpiCypher 14-1048). Library preparation was performed with 5 ng of DNA (or the total amount recovered if less than 5 ng) using the CUTANA™ Library Prep Kit (EpiCypher 14-1001/14-1002). Libraries were run on an Illumina NextSeq2000 with paired-end sequencing (2×50 bp). Sample sequencing depth (vector control cells/overexpressing cells) was 5.9/6.2 million reads (IgG), 13.7/12.6 million reads (H3K4me3), 8.1/5.2 million reads (GATA3), and 5.7/5.9 million reads (HA Tag). Data were aligned to the hg19 genome using Bowtie2. Data were filtered to remove duplicates, multi-aligned reads, and ENCODE DAC Exclusion List regions.
- Type: Polyclonal
- Host: Rabbit
- Applications: CUT&RUN, WB
- Reactivity: HA Epitope (YPYDVPDYA)
- Format: Antigen affinity-purified
- Target Size: N/A
- CUT&RUN: 0.5 µg per reaction
- Western Blot (WB): 1:1,000 – 1:30,000