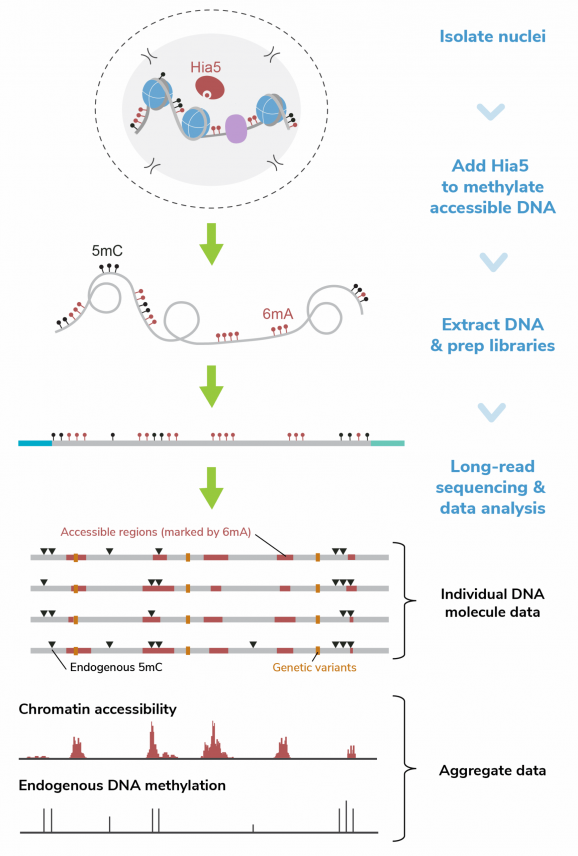
CUTANA™ Hia5 for Fiber-seq
CUTANA™ Hia5 is the key enzyme for Fiber-seq [1], a multiomic long-read sequencing (LRS) assay that simultaneously profiles chromatin accessibility, DNA methylation, and genetic variation at single-molecule resolution. By layering chromatin accessibility on top of standard LRS workflows, Fiber-seq uniquely links epigenetic information (DNA methylation) and complex genetic architecture (e.g., structural variants and haplotype phasing) to chromatin states within the same molecules of DNA.
Hia5 is a DNA adenine methyltransferase (MTase) that uses S-adenosylmethionine (SAM) to catalyze methylation at the N6 position of adenine, producing N6-methyladenine (6mA). Fiber-seq leverages the nonspecific activity of Hia5 to methylate adenines within accessible chromatin regions in situ. This records chromatin accessibility information onto native DNA while preserving endogenous methylation (5mC). Hia5-modified DNA is fully compatible with LRS on both Pacific Biosciences® (PacBio® HiFi Sequencing) and Oxford Nanopore Technologies® (ONT® Nanopore Sequencing) platforms.
Key Advantages
Multiomic insights in one assay – Fiber-seq simultaneously profiles chromatin accessibility, DNA methylation, and genetic variants, consolidating what typically requires three or more separate assays into one long-read sequencing assay.
Highest activity 6mA-MTase – Faster labeling via Hia5 enables higher resolution of accessible DNA, with demonstrated utility in protein footprinting applications to resolve transcription factor binding and nucleosome positioning [2-4].
Access complex genomic regions – Profile chromatin accessibility in repetitive and structurally complex regions such as transposable elements, centromeres, telomeres, segmental duplications, and chromosomal rearrangements.
Easy workflow integration – Fiber-seq can be performed in <2 hours directly upstream of routine long-read sequencing library preparation.
Resolve chromatin state heterogeneity – PCR-free approach leverages long-read sequencing to directly profile individual DNA molecules, uncovering chromatin structure heterogeneity in cell populations often missed by short-read techniques.
Background References:
[1] Stergachis et al. Science (2020). PMID: 32587015
[2] Tullius et al. Mol. Cell (2024). PMID: 39191261
[3] Dubocanin et al. Cell Genom. (2025). PMID: 40147439
[4] Vollger et al. bioRxiv (2024). DOI: 10.1101/2024.06.14.599122
[5] Jha et al. Genome Res. (2024). PMID: 38849157
Figure 1: Fiber-seq methods
Fiber-seq was performed as described in the CUTANA Fiber-seq Protocol v1.0 (see Fiber-seq Protocols page). In brief:
Fiber-seq was performed using 1M K562 nuclei. Nuclei were incubated with Hia5 and co-factor SAM for 10 minutes at 25°C. Genomic DNA was purified, fragmented to 10-15 kb, and prepared for sequencing on a PacBio® Revio® SMRT® Cell. The library was sequenced to achieve >30x coverage. Data were aligned to the hg38 genome using PacBio® SMRT® Link tools, and Fiber-seq specific QC was performed using fibertools [5].
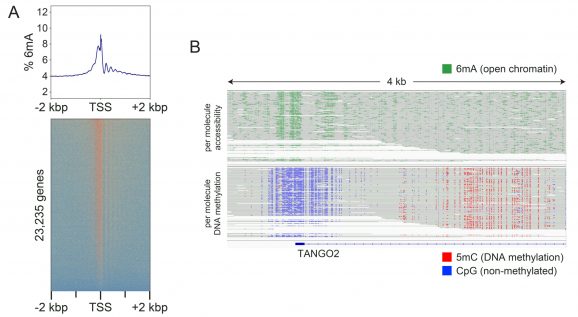
Figure 2: Fiber-seq simultaneously detects open chromatin (6mA) and DNA methylation (5mC)
Fiber-seq was performed as shown in Figure 1.
(A) The Fiber-seq open chromatin signal (6mA) is shown as a heatmap aligned to the transcription start sites (TSS) of 23,235 genes, with the composite signal displayed above. Strong 6mA enrichment is observed at TSSs and within gene bodies, reflecting labeling of accessible DNA at TSSs and linker DNA between nucleosomes in transcriptionally active genes.
(B) A representative locus (TANGO2) illustrates single-molecule patterns of open chromatin (6mA, green; upper track) and endogenous DNA methylation (5mC, red; lower track). Consistent with active transcription, the TANGO2 promoter region displays open chromatin and unmethylated CpGs (blue; lower track) near the TSS, with 5mC observed within the gene body.
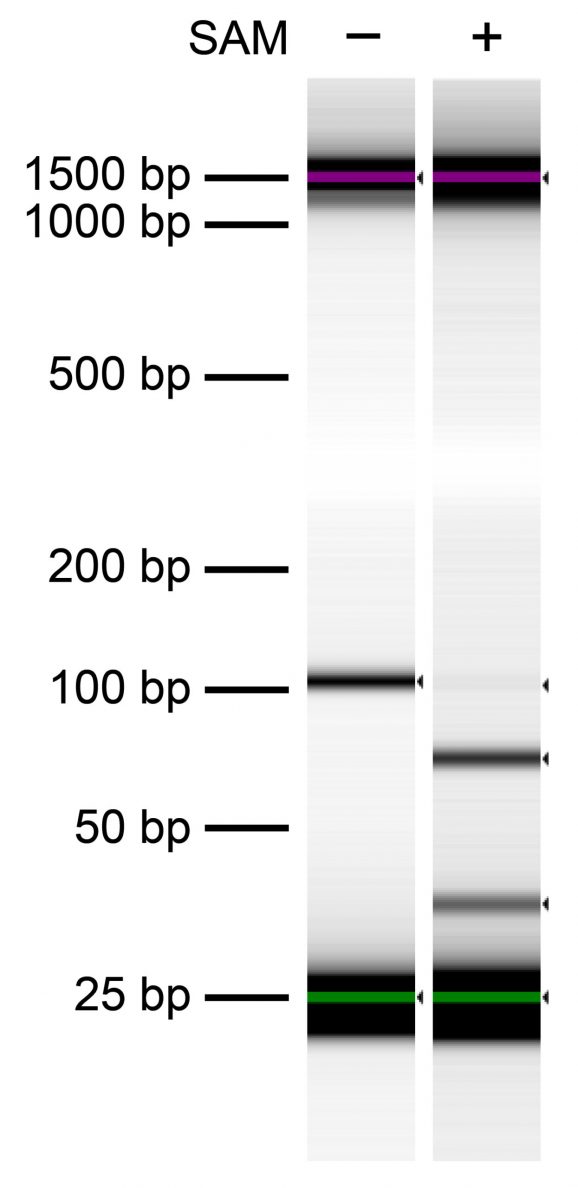
Figure 3: In vitro confirmation of Hia5 6mA methyltransferase activity
Hia5 MTase activity was tested in vitro using a synthetic DNA template (122 bp) containing a single GATC motif. After Hia5 reactions in the presence (+) or absence (-) of the methyl donor SAM, DNA was treated with the methylation-sensitive DpnI restriction enzyme. DNA was then resolved via Agilent TapeStation® D1000 Screen Tape. DpnI-mediated cleavage (81 and 41 bp cut fragments) was observed only in the presence of SAM, confirming Hia5- catalyzed 6mA to produce the DpnI restriction site (G[6mA]TC).
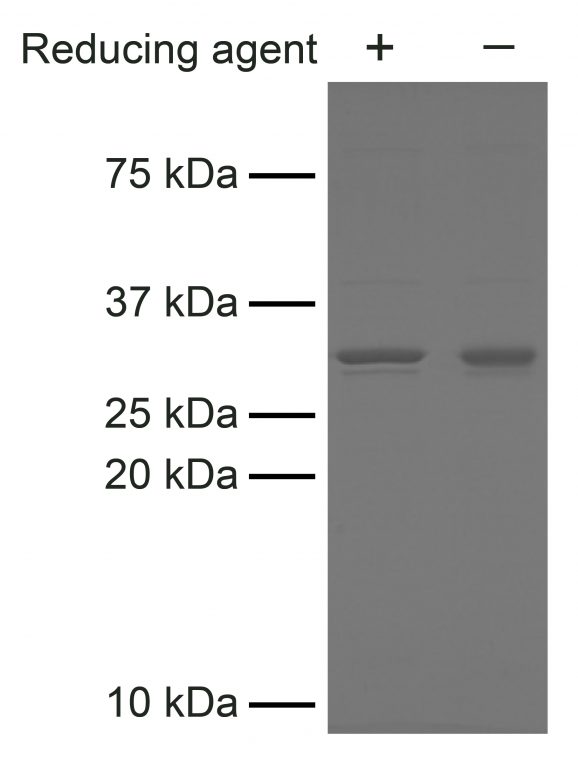
Figure 4: SDS-PAGE analysis of Hia5 purity
Hia5 (2 μg) was resolved on a 15% SDS-PAGE gel and visualized with Coomassie blue staining. Molecular weight standards are shown for reference. Hia5 migrates at the expected size of 33.8 kDa. Samples were run with and without reducing agent to confirm the absence of disulfide bridges, which can contribute to enzyme instability.
- Type: Adenine Methyltransferase
- Mol Wgt: 33.8 kDa
- Host: E. coli
- Epitope Tag: 6xHis
Storage
Stable for six months at -80ºC from date of receipt.
Formulation
50 mM Tris pH 8.0, 1 mM DTT, 250 mM NaCl, 10% glycerol
For detailed applications and uses of this product, see the Fiber-seq protocol. In brief:
In human cells, optimal 6mA labeling is achieved using the recommended reaction conditions below. Optimal 6mA labeling conditions will need to be empirically determined for other organisms (e.g., yeast, plants) or if labeling a different quantity of nuclei.
Recommended usage: Isolate nuclei using CUTANA Nuclei Extraction Buffer (EpiCypher 21-1026), following the nuclei extraction section of the Fiber-seq protocol. Resuspend 1 million human nuclei in 56.5 µL reaction buffer (15 mM Tris pH 8.0, 15 mM NaCl, 60 mM KCl, 1 mM EDTA, 0.5 mM EGTA, 0.5 mM spermidine). Add 2 µL (30X dilution) of the supplied Hia5 enzyme and 1.5 µL 32 mM SAM (60 µL final volume). Incubate reactions for 10 minutes at 25ºC. Quench by adding 6 µL of 10% SDS. Proceed to DNA purification (EpiCypher recommends using NEB® Monarch® Spin gDNA Extraction Kit, NEB #T3010) and LRS library preparation for preferred LRS platform.