H3K4me1 Antibody, SNAP-Certified™ for CUT&RUN and CUT&Tag
19 in stock
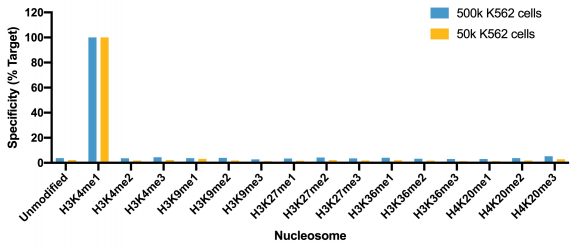
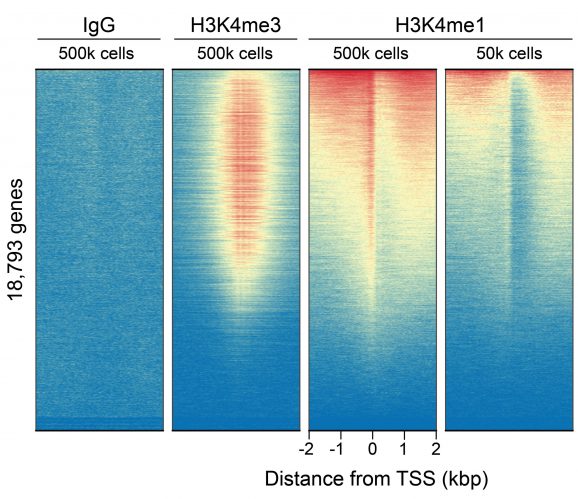
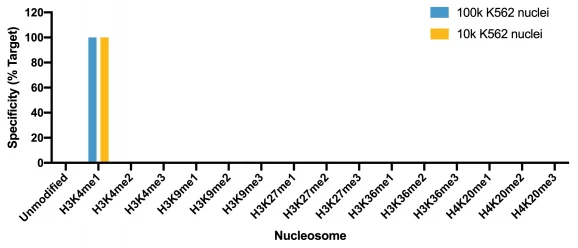
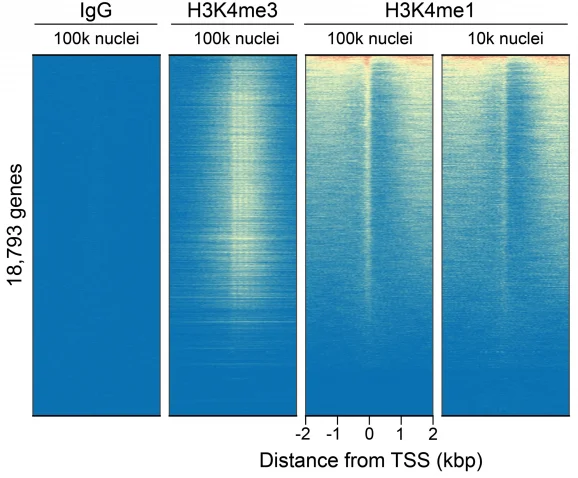
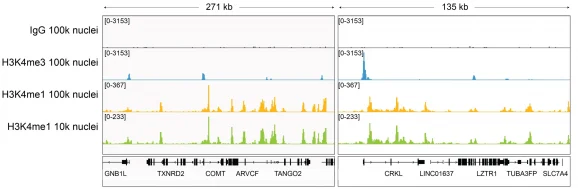
This H3K4me1 (histone H3 lysine 4 monomethyl) antibody meets EpiCypher’s lot-specific SNAP-Certified™ criteria for specificity and efficient target enrichment in both CUT&RUN and CUT&Tag applications. This requires <20% cross-reactivity to related histone PTMs determined using the SNAP-CUTANA™ K-MetStat Panel of spike-in controls (EpiCypher 19-1002, Figures 1 and 4). High target efficiency is confirmed by consistent genomic enrichment at varying cell inputs: 500k and 50k cells in CUT&RUN (Figures 2-3); 100k and 10k cells in CUT&Tag (Figures 5-6). High efficiency antibodies display similar peak structures at representative loci (Figures 3 and 6) and highly conserved genome-wide signal (Figures 2 and 5) even at reduced cell numbers. H3K4me1 either flanks H3K4me3 at the transcription start site (TSS) or coincides with H3K4me3 (Figures 2-3, 5-6) [Bae & Lesch, 2020].
Background References:
[1] Bae & Lesch Front Cell Dev. Biol. (2020). PMID: 32432110
Figure 1: SNAP specificity analysis in CUT&RUN
Figure 2: CUT&RUN genome-wide enrichment
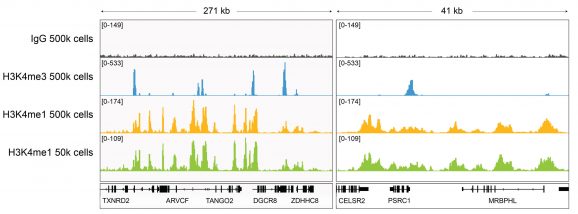
Figure 3: H3K4me1 CUT&RUN representative browser tracks
CUT&RUN was performed as described above. Gene browser shots were generated using the Integrative Genomics Viewer (IGV, Broad Institute). H3K4me1 antibody tracks display the characteristic enrichment known to be consistent with the function of this PTM [Bae & Lesch, 2020]. Similar results in peak structure and location were observed for both 500k and 50k cell inputs.
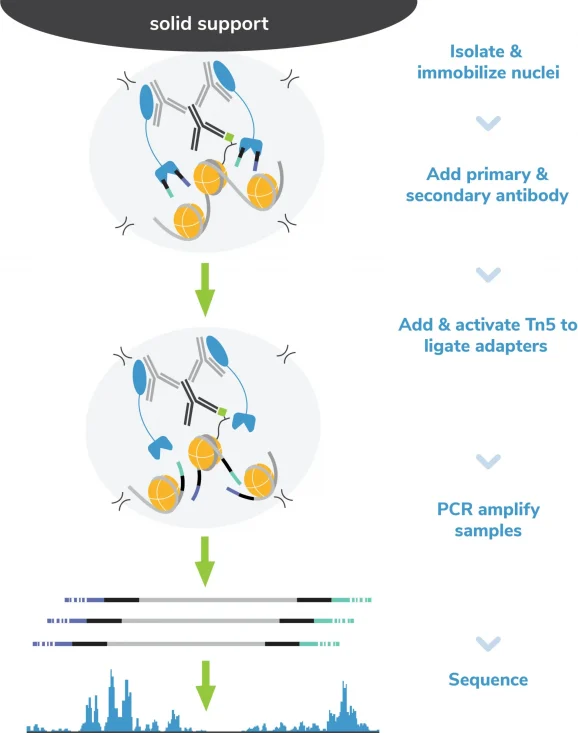
CUT&RUN methods
CUT&RUN was performed on 500k and 50k K562 cells with the SNAP-CUTANA™ K-MetStat Panel (EpiCypher 19-1002) spiked-in prior to the addition of 0.5 µg of either IgG negative control (EpiCypher 13-0042), H3K4me3 positive control (EpiCypher 13-0041), or H3K4me1 antibodies. The experiment was performed using the CUTANA™ ChIC/CUT&RUN Kit v3.0 (EpiCypher 14-1048). Library preparation was performed with 5 ng of CUT&RUN enriched DNA (or the total amount recovered if less than 5 ng) using the CUTANA™ CUT&RUN Library Prep Kit (EpiCypher 14-1001/14-1002). Both kit protocols were adapted for high throughput Tecan liquid handling. Libraries were run on an Illumina NextSeq2000 with paired-end sequencing (2×50 bp). Sample sequencing depth was 6.7 million reads (IgG 50k cell input), 11.5 million reads (IgG 500k cell input), 10.2 million reads (H3K4me3 50k cell input) and 16.7 million reads (H3K4me3 500k cell input). Data were aligned to the hg19 genome using Bowtie2. Data were filtered to remove duplicates, multi-aligned reads, and ENCODE DAC Exclusion List regions.
Figure 4: SNAP specificity analysis in CUT&Tag
CUT&Tag was performed as described above. CUT&Tag sequencing reads were aligned to the unique DNA barcodes corresponding to each nucleosome in the K-MetStat panel (x-axis). Data are expressed as a percent relative to on-target recovery (H3K4me1 set to 100%).
Figure 5: CUT&Tag genome-wide enrichment
Figure 6: H3K4me1 CUT&Tag representative browser tracks
CUT&Tag was performed as described above. Gene browser shots were generated using the Integrative Genomics Viewer (IGV, Broad Institute). H3K4me1 antibody tracks display the characteristic enrichment known to be consistent with the function of this PTM [Bae & Lesch, 2020]. Similar results in peak structure and location were observed for both 100k and 10k nuclei inputs.
CUT&Tag methods
CUT&Tag was performed on 100k and 10k K562 nuclei with the SNAP-CUTANA™ K-MetStat Panel (EpiCypher 19-1002) spiked-in prior to the addition of 0.5 µg of either IgG negative control (EpiCypher 13-0042), H3K4me3 positive control (EpiCypher 13-0041), or H3K4me1 antibodies. The experiment was performed using the CUTANA™ CUT&Tag Kit v1 (EpiCypher 14-1102). Libraries were run on an Illumina NextSeq2000 with paired-end sequencing (2×50 bp). Sample sequencing depth was 16.8 million reads (IgG 500k cell input), 14.4 million reads (H3K4me3 500k cell input), 26.2 million reads (H3K4me1 500k cell input) and 11.4 million reads (H3K4me1 50k cell input). Data were aligned to the hg19 genome using Bowtie2. Data were filtered to remove duplicates, multi-aligned reads, and ENCODE DAC Exclusion List regions.
- Type: Monoclonal [2088-1F4]
- Host: Rabbit
- Applications: CUT&RUN, CUT&Tag
- Reactivity: Human, Wide Range (Predicted)
- Format: Protein A affinity-purified
- Target Size: 15 kDa
Protein A affinity-purified recombinant monoclonal antibody in Borate buffered saline pH 8.0, 0.09% sodium azide
- CUT&RUN: 0.5 µg per reaction
- CUT&Tag: 0.5 µg per reaction
- UniProt ID: H3.1 – P68431
- Alternate Names: H3, H3/a, H3/b, H3/c, H3/d