Why are designer nucleosomes so important for epigenetics research?
Nucleosome substrates are emerging tools for the in vitro study of chromatin epigenetic regulation, with applications from basic chromatin biology research to high-throughput drug screening. The advent of recombinant designer nucleosomes (dNucs) carrying fully defined covalent modifications of the histone proteins (e.g. lysine acylation) or wrapping DNA (e.g. 5’ methylcytosine) enables novel or dramatically improved approaches (e.g. reader binding, enzymatic assays, antibody profiling) for next generation chromatin research.
In this post we are going to talk about the minimal thresholds for the assembly and quality validation of these reagents.
Recombinant nucleosomes as physiological substrates for drug discovery and drug development
Here at EpyCypher, we have pioneered the commercial development of recombinant nucleosomes for next generation epigenetic assays and tool development (see Table 1).
Mounting evidence demonstrates that nucleosomes are the optimal substrates to characterize many chromatin regulators [1-4]. For example, NSD2 methyltransferase (associated with oncogenic reprogramming in multiple myeloma [5, 6]) requires nucleosomal substrates for in vitro activity [7].
In the following table, there is a list of all the recombinant nucleosomes available from EpiCypher.
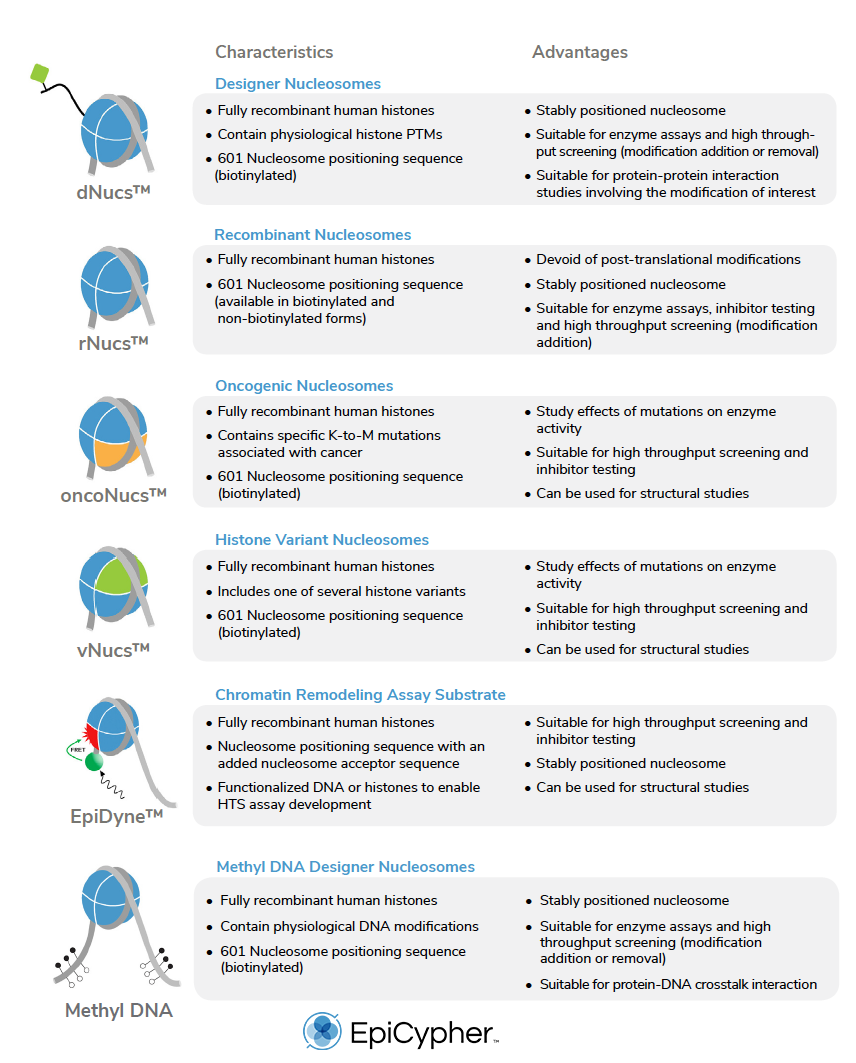
Nucleosomes carrying defined covalent modifications (termed designer nucleosomes or ‘dNucs’) also provide excellent substrates for a range of reader and eraser assays. For instance, binding of the BRD4 tandem bromodomain is significantly increased when lysine residues on both histone H3 and H4 are acetylated [4], while LSD1 demethylase and the SIRTUIN family of histone deacetylases require acetylated nucleosomes to mimic in vivo target selectivity [9-13].
In addition to assays that examine the addition, recognition or removal of histone modifications, recombinant nucleosomes can also be made compatible with chromatin remodeling assays (see EpiDyne™ product family in Table 1).
Of note, the SWI/SNF family of chromatin remodeling complexes are emerging as high value therapeutic targets, with research indicating the need for nucleosome substrates to accurately interrogate the ATPase-dependent function of these enzymes.
REFERENCES
1 Kim, J., et al., The n-SET domain of Set1 regulates H2B ubiquitylation-dependent H3K4 methylation. Mol Cell, 2013. 49(6): p. 1121-33.
2. Whitcomb, S.J., et al., Histone monoubiquitylation position determines specificity and direction of enzymatic cross-talk with histone methyltransferases Dot1L and PRC2. J Biol Chem, 2012. 287(28): p. 23718-25.
3. McGinty, R.K., et al., Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature, 2008. 453(7196): p. 812-6.
4. Nguyen, U.T., et al., Accelerated chromatin biochemistry using DNA-barcoded nucleosome libraries.
Nat Methods, 2014. 11(8): p. 834-40.
5. Keats, J.J., et al., Overexpression of transcripts originating from the MMSET locus characterizes all t(4;14) (p16;q32)-positive multiple myeloma patients. Blood, 2005. 105(10): p. 4060-9.
6. Lauring, J., et al., The multiple myeloma associated MMSET gene contributes to cellular adhesion, clonogenic growth, and tumorigenicity. Blood, 2008. 111(2): p. 856-64.
7. Li, Y., et al., The target of the NSD family of histone lysine methyltransferases depends on the nature of the substrate. J Biol Chem, 2009. 284(49): p. 34283-95.
8. Strelow, J.M., et al., The Use of Nucleosome Substrates Improves Binding of SAM Analogs to SETD8. J Biomol Screen, 2016. 21(8): p. 786-94.
9. Shi, Y.J., et al., Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell, 2005. 19(6): p. 857-64.
10. Yang, M., et al., Structural basis for CoREST-dependent demethylation of nucleosomes by the human LSD1 histone demethylase. Mol Cell, 2006. 23(3): p. 377-87.
