SKU: 13-0028
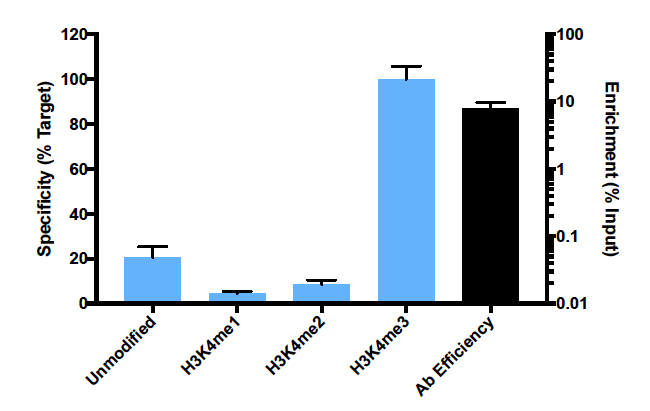
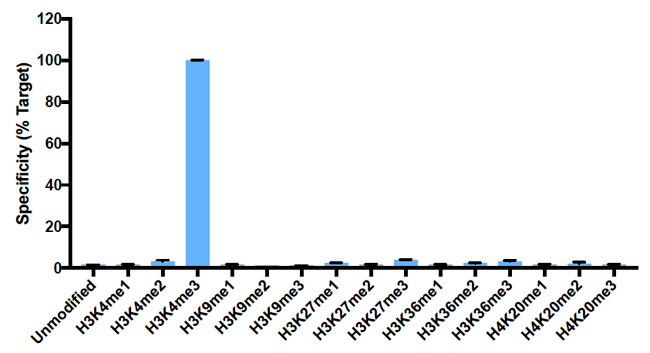
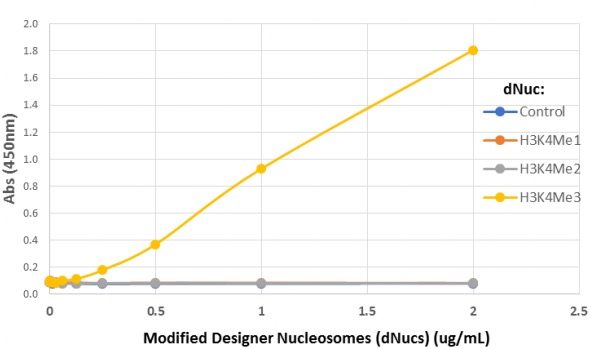
{"url":"https://www.epicypher.com/products/antibodies/snap-chip-certified-antibodies/histone-h3k4me3-antibody-snap-chip-certified","add_this":[{"service":"facebook","annotation":""},{"service":"email","annotation":""},{"service":"print","annotation":""},{"service":"twitter","annotation":""},{"service":"linkedin","annotation":""}],"warranty":"","gtin":"","max_purchase_quantity":0,"options":[],"id":"626","can_purchase":false,"meta_description":"Histone H3K4me3 antibody. This antibody meets EpiCypher’s “SNAP-ChIP® Certified” criteria for specificity and efficient target enrichment in a ChIP experiment.","category":[],"meta_keywords":"h3k4me3 antibody","AddThisServiceButtonMeta":"","images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/626/578/Screen_Shot_2018-09-25_at_2.00.27_PM__52796.1538135192.png?c=2","alt":"Histone H3K4me3 Antibody, SNAP-ChIP Certified *DISCONTINUED*"}],"main_image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/626/578/Screen_Shot_2018-09-25_at_2.00.27_PM__52796.1538135192.png?c=2","alt":"Histone H3K4me3 Antibody, SNAP-ChIP Certified *DISCONTINUED*"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=626","shipping":[],"num_reviews":0,"weight":"0.00 LBS","custom_fields":[{"id":"452","name":"Pack Size","value":"100 μg"}],"sku":"13-0028","description":"<table cellpadding=\"2\" cellspacing=\"2\" style=\"width: 100%\">\n <tbody>\n <tr valign=\"top\">\n <a\n style=\"color: #fff\"\n href=\"/products/antibodies/h3k4me3-antibody-snap-certified-for-cut-run-and-cut-tag\"\n >\n <p\n style=\"\n background-color: #4698cb;\n color: #fff;\n padding: 1.3rem;\n text-align: center;\n border-radius: 12px;\n margin-top: 2.5rem;\n \"\n >\n Product Discontinued - See 13-0060 as a recommended alternative\n </p>\n </a>\n <td>\n <table cellpadding=\"1\" cellspacing=\"1\" style=\"width: 95%\">\n <tbody>\n <tr>\n <td>\n <table cellpadding=\"0\" cellspacing=\"0\" style=\"width: 100%\">\n <tbody>\n <tr>\n <td valign=\"top\">\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><strong\n >T<span style=\"font-size: 12pt\"\n >ype: </span\n ></strong\n ><span style=\"font-size: 12pt\">Monoclonal</span></span\n >\n </td>\n <td valign=\"top\">\n <span style=\"font-size: 12pt\"\n ><span\n style=\"font-family: arial, helvetica, sans-serif\"\n ><strong>Host: </strong>Rabbit</span\n ></span\n >\n </td>\n </tr>\n <tr>\n <td valign=\"top\">\n <span style=\"font-size: 12pt\"\n ><span\n style=\"font-family: arial, helvetica, sans-serif\"\n ><strong>Mol Wgt.: </strong>15 kDa</span\n ></span\n >\n </td>\n <td valign=\"top\">\n <span style=\"font-size: 12pt\"\n ><span\n style=\"font-family: arial, helvetica, sans-serif\"\n ><strong>Appl.: </strong>ChIP, ChIP-Seq,\n ELISA</span\n ></span\n >\n </td>\n </tr>\n <tr>\n <td valign=\"top\">\n <span style=\"font-size: 12pt\"\n ><span\n style=\"font-family: arial, helvetica, sans-serif\"\n ><strong>Format: </strong>Aff. Pur.\n IgG</span\n ></span\n >\n </td>\n <td valign=\"top\">\n <span style=\"font-size: 12pt\"\n ><span\n style=\"font-family: arial, helvetica, sans-serif\"\n ><strong>Reactivity: </strong>H, M, WR</span\n ></span\n >\n </td>\n </tr>\n </tbody>\n </table>\n </td>\n </tr>\n <tr>\n <td valign=\"top\">\n <span style=\"font-size: 12pt\"\n ><span style=\"font-family: arial, helvetica, sans-serif\"\n ><strong\n >Histone H3K4me3 Antibody - SNAP-ChIP® Certified\n Description:</strong\n >\n This antibody meets EpiCypher’s “<a\n href=\"https://www.epicypher.com/technologies/snap-chip-spike-ins/certified-antibodies\"\n >SNAP-ChIP<sup>®</sup> Certified</a\n >” criteria for specificity and efficient target\n enrichment in a ChIP experiment (<20% cross-reactivity\n across the panel, >5% recovery of target input). Histone\n H3 is one of the four proteins that are present in the\n nucleosome, the basic repeating subunit of chromatin,\n consisting of 147 base pairs of DNA wrapped around an\n octamer of core histone proteins (H2A, H2B, H3 and H4). This\n antibody reacts to H3K4me3 and no cross reactivity with\n H3K4me1 or H3K4me2, or other lysine methylations in the\n EpiCypher SNAP-ChIP K-MetStat panel, is detected.\n </span></span\n ><br />\n </td>\n </tr>\n <tr>\n <td valign=\"top\">\n <span style=\"font-size: 12pt\"\n ><span style=\"font-family: arial, helvetica, sans-serif\"\n ><strong\n >Histone H3K4me3 Antibody - SNAP ChIP Certified\n Immunogen:</strong\n >\n A synthetic peptide corresponding to histone H3\n trimethylated at lysine 4.</span\n ></span\n >\n <p></p>\n </td>\n </tr>\n <tr>\n <td valign=\"top\">\n <span style=\"font-size: 12pt\"\n ><span style=\"font-family: arial, helvetica, sans-serif\"\n ><strong\n >Histone H3K4me3 Antibody - SNAP ChIP Certified\n Formulation:</strong\n >\n Protein A affinity-purified antibody (1 mg/mL) in PBS, with\n 0.09% sodium azide, 1% BSA, and 50% glycerol.</span\n ></span\n >\n <p></p>\n </td>\n </tr>\n <tr>\n <td valign=\"top\">\n <span style=\"font-size: 12pt\"\n ><span style=\"font-family: arial, helvetica, sans-serif\"\n ><strong\n >Histone H3K4me3 Antibody - SNAP ChIP Certified Storage\n and Stability:</strong\n >\n Stable for 1 years at -20°C from date of receipt.</span\n ></span\n >\n <p></p>\n </td>\n </tr>\n <tr>\n <td valign=\"top\">\n <span style=\"font-size: 12pt\"\n ><span style=\"font-family: arial, helvetica, sans-serif\"\n ><strong\n >Histone H3K4me3 Antibody - SNAP ChIP Certified\n Application Notes</strong\n >\n <span style=\"font-weight: normal\"\n ><strong>Recommended dilutions:</strong><br />\n ChIP 2 - 5 μg per 1x10<sup>6</sup> cells ELISA 1 - 10\n μg/mL</span\n ></span\n ></span\n >\n </td>\n </tr>\n <tr>\n <td valign=\"top\">\n <span style=\"font-size: 12pt\"\n ><span style=\"font-family: arial, helvetica, sans-serif\"\n ><strong>Background References:</strong></span\n ></span\n ><br />\n\n <span style=\"font-size: 12pt\"\n ><span style=\"font-family: arial, helvetica, sans-serif\"\n >Grzybowski A et al. 2015 Mol Cell 58: 886-899 PMID\n <a\n href=\"https://www.ncbi.nlm.nih.gov/pubmed/26004229\"\n target=\"_new\"\n title=\"Calibrating ChIP-Seq with Nucleosomal Internal Standards to Measure Histone Modification Density Genome Wide\"\n >26004229</a\n ></span\n ></span\n ><br />\n <span style=\"font-size: 12pt\"\n ><span style=\"font-family: arial, helvetica, sans-serif\">\n <br />\n <strong>Product References:</strong><br />\n Shah RN et al. 2018 A et al. 2015 Mol Cell 72: 162 PMID\n <a\n href=\"https://www.ncbi.nlm.nih.gov/pubmed/30244833\"\n target=\"_new\"\n title=\"Examining the Roles of H3K4 Methylation States with Systematically Characterized Antibodies\"\n >30244833</a\n >\n <br />\n Lam K-WG et al. 2019 Nat. Commun. 10: 3821 PMID\n <a\n href=\"https://www.ncbi.nlm.nih.gov/pubmed/31444359\"\n target=\"_new\"\n title=\"Examining the Roles of H3K4 Methylation States with Systematically Characterized Antibodies\"\n >31444359</a\n >\n <br /> </span\n ></span>\n\n <p></p>\n </td>\n </tr>\n <tr>\n <td valign=\"top\">\n <span style=\"font-size: 12pt\"\n ><span style=\"font-family: arial, helvetica, sans-serif\"\n >View technical datasheet for this product.\n <a\n href=\"https://www.epicypher.com/content/documents/tds/13-0028.pdf\"\n target=\"_new\"\n >\n <img\n alt=\"13-0028 Datasheet\"\n height=\"40\"\n src=\"https://cdn7.bigcommerce.com/s-y9o92/content/documents/tds/icon.png\"\n width=\"30\" /></a></span\n ></span>\n </td>\n </tr>\n <tr>\n <td>\n <p></p>\n\n <p>\n <span style=\"font-size: 10pt\"\n ><span style=\"font-family: arial, helvetica, sans-serif\"\n ><strong>Applications Key: ChIP</strong>-Chromatin\n IP; <strong>E</strong>-ELISA; <strong>FACS</strong>-Flow\n cytometry; <strong>IF</strong>-Immunofluorescence;\n <strong>IHC</strong>-Immunohistochemistry;\n <strong>IP</strong>-Immunoprecipitation;\n <strong>WB</strong>-Western Blotting</span\n ></span\n >\n </p>\n\n <p>\n <span style=\"font-size: 10pt\"\n ><span style=\"font-family: arial, helvetica, sans-serif\"\n ><strong>Reactivity Key: B</strong>-Bovine;\n <strong>Ce</strong>-C. elegans;\n <strong>Ch</strong>-Chicken; <strong>Dm</strong>-\n Drosophila; <strong>Eu</strong>-Eukaryote;\n <strong>H</strong>-Human; <strong>M</strong>-Mouse;\n <strong>Ma</strong>-Mammal; <strong>R</strong>-Rat;\n <strong>Sc</strong>-S.cerevesiae; <strong>Sp</strong>-S.\n pombe; <strong>WR</strong>-Wide Range (predicted);\n <strong>X</strong>-Xenopus;\n <strong>Z</strong>-Zebrafish</span\n ></span\n >\n </p>\n </td>\n </tr>\n </tbody>\n </table>\n </td>\n <td>\n <table cellpadding=\"3\" cellspacing=\"3\" style=\"width: 300px\">\n <tbody>\n <tr>\n <td>\n <span style=\"font-family: arial, helvetica, sans-serif\">\n <a\n href=\"https://cdn11.bigcommerce.com/s-y9o92/product_images/uploaded_images/h3k4me3-epicypher-13-0028.jpg?t=1600899553&_ga=2.200012717.241922123.1600715432-402741698.1598987696\"\n target=\"_new\"\n ><img\n alt=\"13-0028 SNAP-ChIP Data\"\n height=\"100\"\n src=\"https://cdn11.bigcommerce.com/s-y9o92/product_images/uploaded_images/h3k4me3-epicypher-13-0028.jpg?t=1600899553&_ga=2.200012717.241922123.1600715432-402741698.1598987696\"\n width=\"150\" /></a\n ></span>\n </td>\n </tr>\n <tr>\n <td valign=\"top\">\n <p>\n <span style=\"font-size: 11pt\"\n ><span style=\"font-family: arial, helvetica, sans-serif\"\n ><strong>Representative SNAP-ChIP-seq results: </strong>\n Cumulative histogram plot and heatmap of signal intensity\n depict H3K4me3 ChIP-seq data aligned to annotated\n transcription start sites (TSS, +/- 3.0 kb; left). Two\n representative genomic regions depicting H3K4me3 peak\n structure and distribution are shown (right). Data shown\n are representative of H3K4me3 ChIP antibody (EpiCypher\n Catalog No. 13-0028) and are not lot-specific. Native\n ChIP-seq was performed using K562 cells as described (<a\n href=\"https://pubmed.ncbi.nlm.nih.gov/30244833/\"\n target=\"_blank\"\n >Shah et al., Mol Cell 2018</a\n >) with SNAP-ChIP<sup>TM</sup> K-MetStat Spike-in (Catalog\n No.\n <a\n href=\"https://www.epicypher.com/products/nucleosomes/snap-chip-k-metstat-panel\"\n target=\"_blank\"\n >19-1001</a\n >) nucleosome controls added prior to chromatin digestion\n to confirm antibody specificity and ChIP efficiency.\n Paired-end sequencing libraries were prepared using the\n NEBNext<sup>®</sup> Ultra<sup>TM</sup> II DNA Library Prep\n Kit for Illumina<sup>®</sup>. ChIP libraries were\n sequenced on an Illumina<sup>®</sup> NextSeq. Sequencing\n reads were aligned to the human genome using Bowtie 2\n (Johns Hopkins University). Bigwig files of read\n enrichment in binned genomic regions (signal intensity)\n flanking the indicated gene features were used to create a\n cumulative histogram plot and heatmap of signal intensity\n (<a href=\"https://basepairtech.com/\" target=\"_blank\">\n www.basepairtech.com</a\n >). Gene browser shots were generated using the\n Integrative Genomics Viewer (IGV, Broad Institute) with\n the window size denoted (top).\n\n <br />\n <strong>(Click image to enlarge) </strong></span\n ></span\n >\n </p>\n\n <p></p>\n </td>\n </tr>\n\n <tr>\n <td>\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><a\n href=\"https://www.epicypher.com/content/images/products/antibodies/13_0028_qPCR_tds.jpg\"\n target=\"_new\"\n ><img\n alt=\"13-0028 SNAP-ChIP Data\"\n height=\"100\"\n src=\"https://cdn11.bigcommerce.com/s-y9o92/content/images/products/antibodies/13_0028_qPCR_tds.jpg\"\n width=\"150\" /></a\n ></span>\n </td>\n </tr>\n <tr>\n <td valign=\"top\">\n <p>\n <span style=\"font-size: 11pt\"\n ><span style=\"font-family: arial, helvetica, sans-serif\"\n ><strong>SNAP-ChIP qPCR Data: </strong>Histone H3K4me3\n antibody (3 μg) was tested in a native ChIP experiment\n with chromatin from HEK-293 cells (~1x10<sup>6</sup>\n cells) with the SNAP-ChIP K-MetStat Panel (EpiCypher\n Catalog No.\n <a\n href=\"https://www.epicypher.com/snap-chip-k-metstat-panel/\"\n target=\"_new\"\n >19-1001</a\n >). spiked-in prior to micrococcal nuclease digestion.\n Specificity (left Y-axis) was determined by qPCR for the\n DNA barcodes corresponding to modified nucleosomes in the\n SNAP-ChIP panel (x-axis). Black bar represents antibody\n efficiency (right y-axis; log scale) and indicates\n percentage of the target immunoprecipitated relative to\n input. Error bars represent mean ± SEM in replicate\n ChIP experiments.<br />\n <strong>(Click image to enlarge) </strong></span\n ></span\n >\n </p>\n\n <p></p>\n </td>\n </tr>\n <tr>\n <td>\n <span style=\"font-size: 11pt\"\n ><span style=\"font-family: arial, helvetica, sans-serif\"\n ><a\n href=\"https://www.epicypher.com/content/images/products/antibodies/13_0028_ChIP_seq_tds.jpg\"\n target=\"_new\"\n ><img\n alt=\"13-0028 SNAP-ChIP Data\"\n height=\"100\"\n src=\"https://cdn11.bigcommerce.com/s-y9o92/content/images/products/antibodies/13_0028_ChIP_seq_tds.jpg\"\n width=\"150\" /></a></span\n ></span>\n </td>\n </tr>\n <tr>\n <td valign=\"top\">\n <p>\n <span style=\"font-size: 11pt\"\n ><span style=\"font-family: arial, helvetica, sans-serif\"\n ><strong>SNAP-ChIP-seq Data: </strong>Histone H3K4me3\n antibody (5 μg) was tested in a native ChIP experiment\n using chromatin from K-562 cells (~1x10<sup>6</sup> cells)\n with the SNAP-ChIP K-MetStat Panel (~1x10<sup>6</sup>\n cells) with the SNAP-ChIP K-MetStat Panel (EpiCypher\n Catalog No. 19-1001) spiked-in prior to micrococcal\n nuclease digestion. Ten nanograms ChIP DNA was subjected\n to library preparation using the NEBNext® Ultra<sup\n >TM</sup\n >\n II DNA Library Prep Kit for Illumina®. ChIP libraries\n were analyzed by 2x150bp paired end sequencing on an\n Illumina HiSeq 4000. Paired reads were aligned to the\n SNAP-ChIP barcodes using the alignment algorithm available\n at https://www.basepairtech.com/. Specificity was\n determined by normalizing the read counts for each\n barcoded nucleosome IP to the corresponding Input and\n expressing the data as a percent of the nucleosome\n containing the target PTM (% Target). Error bars represent\n mean ± SEM for the duplicate DNA barcodes in a\n single ChIP experiment.<br />\n <strong>(Click image to enlarge) </strong></span\n ></span\n >\n </p>\n\n <p></p>\n </td>\n </tr>\n <tr valign=\"top\">\n <td valign=\"top\">\n <span style=\"font-size: 11pt\"\n ><span style=\"font-family: arial, helvetica, sans-serif\"\n ><a\n href=\"https://www.epicypher.com/content/images/products/antibodies/13_0028_ELISA_tds.jpg\"\n target=\"_new\"\n ><img\n alt=\"13-0028 ELISA\"\n height=\"100\"\n src=\"https://cdn11.bigcommerce.com/s-y9o92/content/images/products/antibodies/13_0028_ELISA_tds.jpg\"\n width=\"127\" /></a></span\n ></span>\n </td>\n </tr>\n <tr>\n <td valign=\"top\">\n <p>\n <span style=\"font-size: 11pt\"\n ><span style=\"font-family: arial, helvetica, sans-serif\"\n ><strong>ELISA Data: </strong>Designer Nucleosomes (dNucs)\n containing the indicated H3K4 modification were used in an\n ELISA assay with Histone H3K4me3 Antibody (10\n μg/mL).<br />\n <strong>(Click image to enlarge) </strong></span\n ></span\n >\n </p>\n\n <p></p>\n </td>\n </tr>\n </tbody>\n </table>\n </td>\n </tr>\n </tbody>\n</table>\n<style>\n .button-group {\n display: none;\n }\n</style>\n","tags":[],"detail_messages":"","availability":"","page_title":"Histone H3K4me3 Antibody | SNAP-ChIP Certified","mpn":"","upc":null,"shipping_messages":[],"rating":0,"title":"Histone H3K4me3 Antibody, SNAP-ChIP Certified *DISCONTINUED*","gift_wrapping_available":false,"min_purchase_quantity":0}
Pack Size: 100 μg