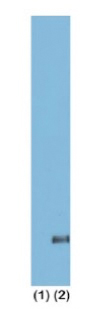
Histone H3K36me3 Antibody, SNAP-ChIP® Certified, CUTANA™ CUT&RUN Compatible *DISCONTINUED*
SKU: 13-0031
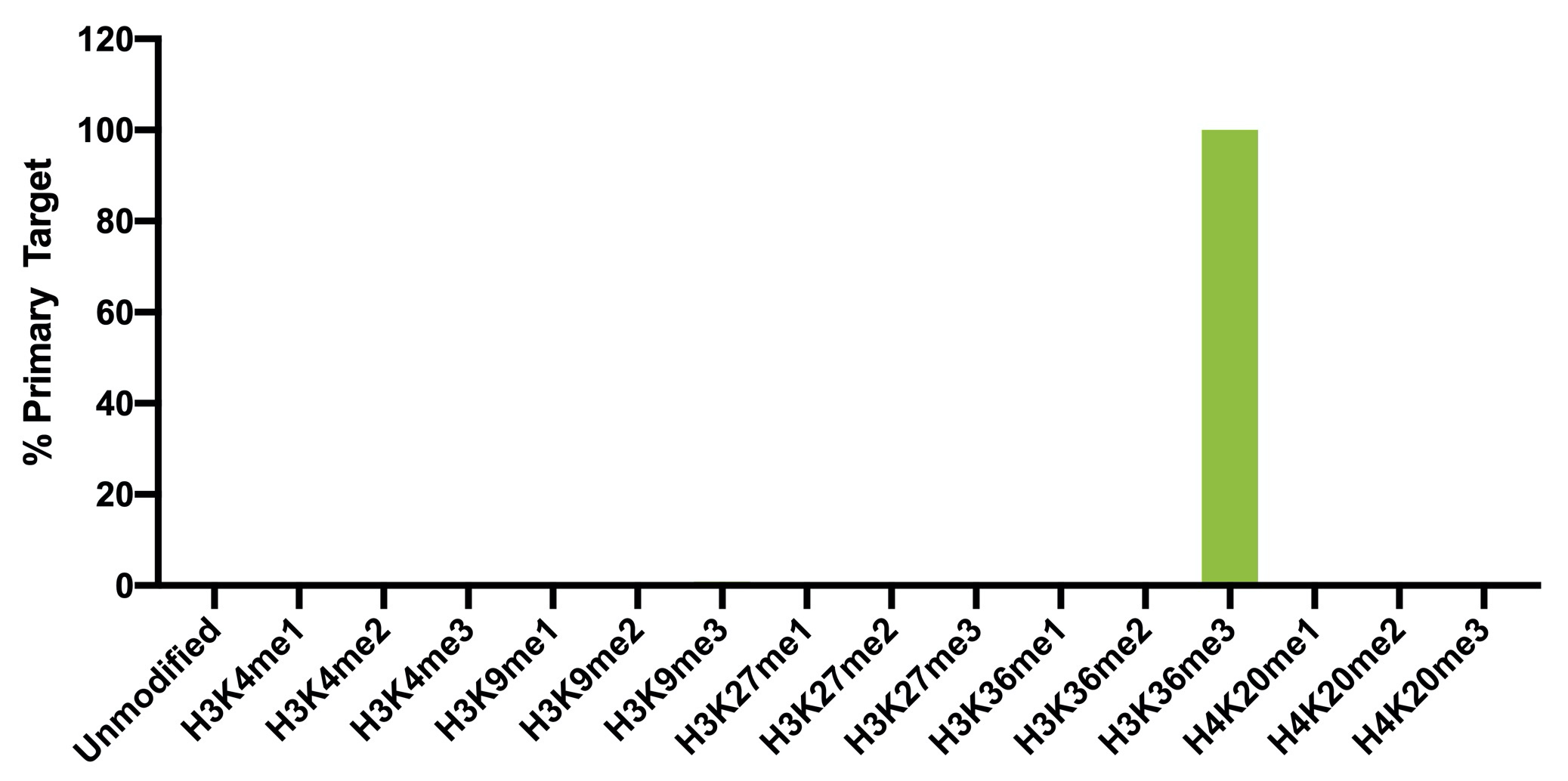
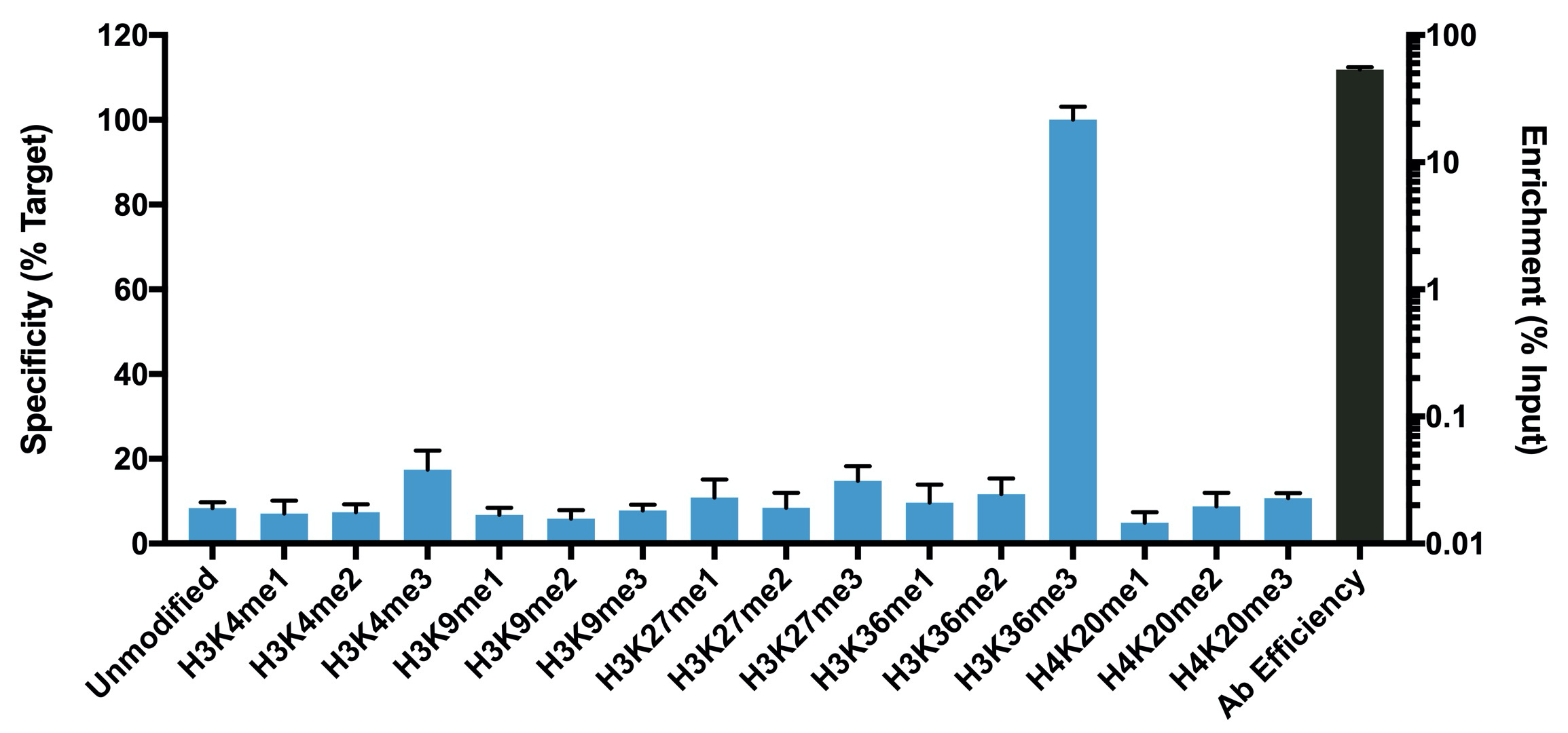
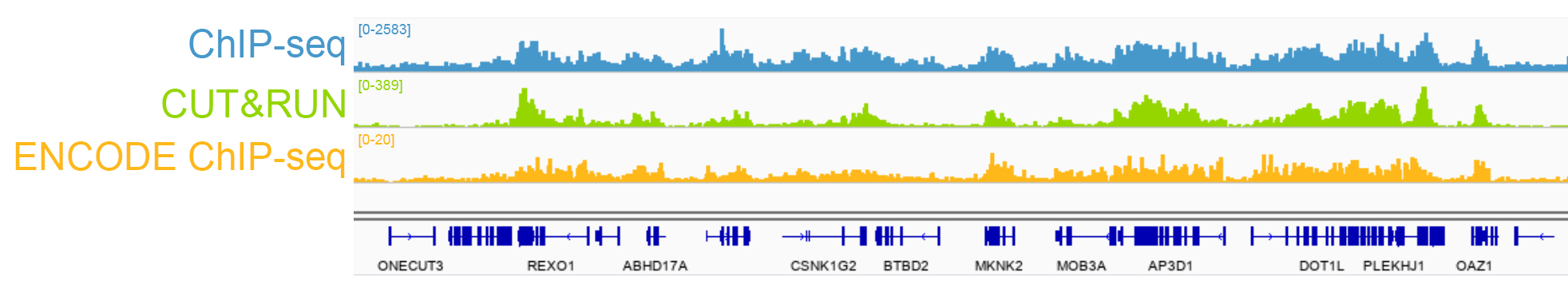
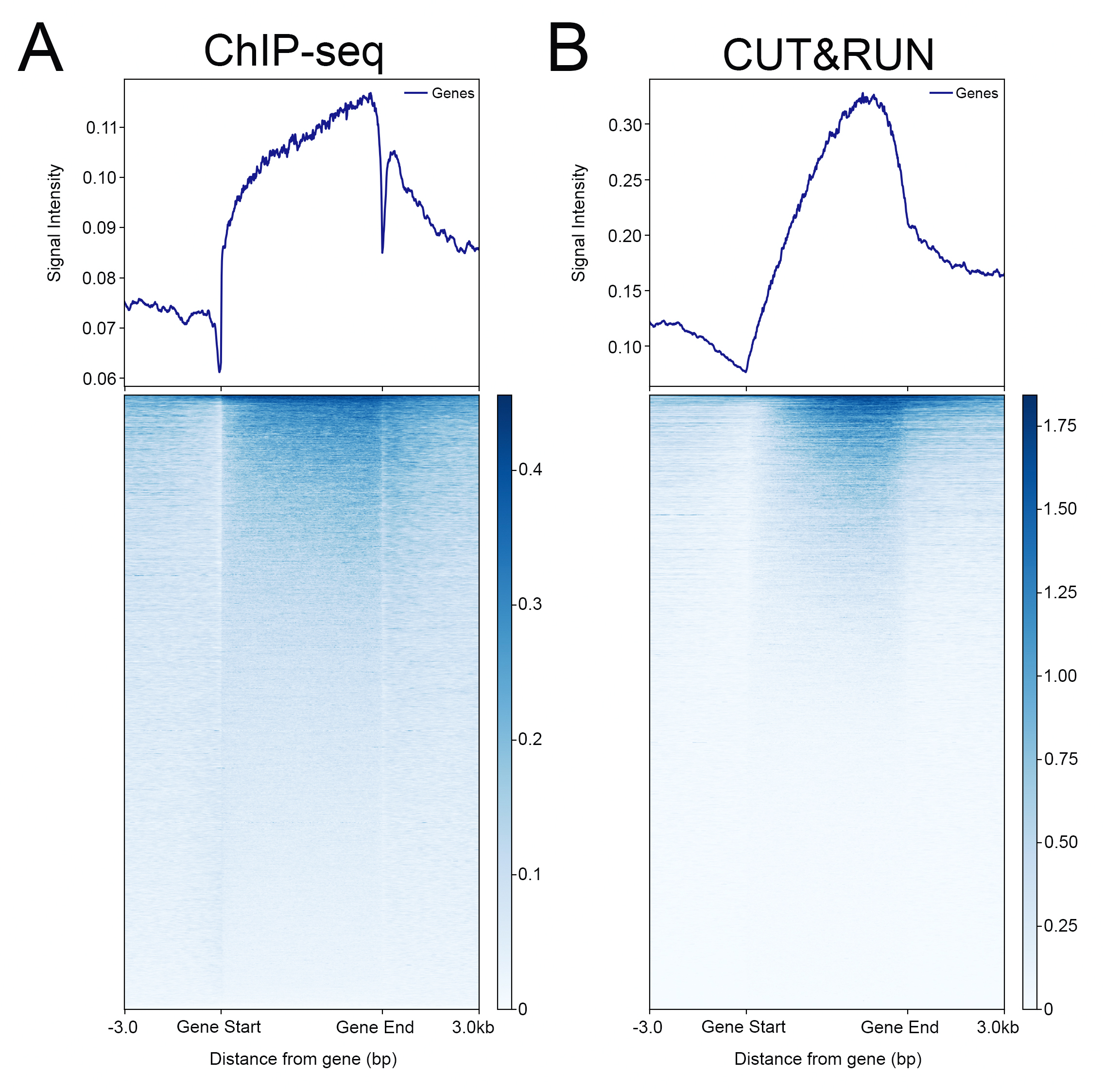
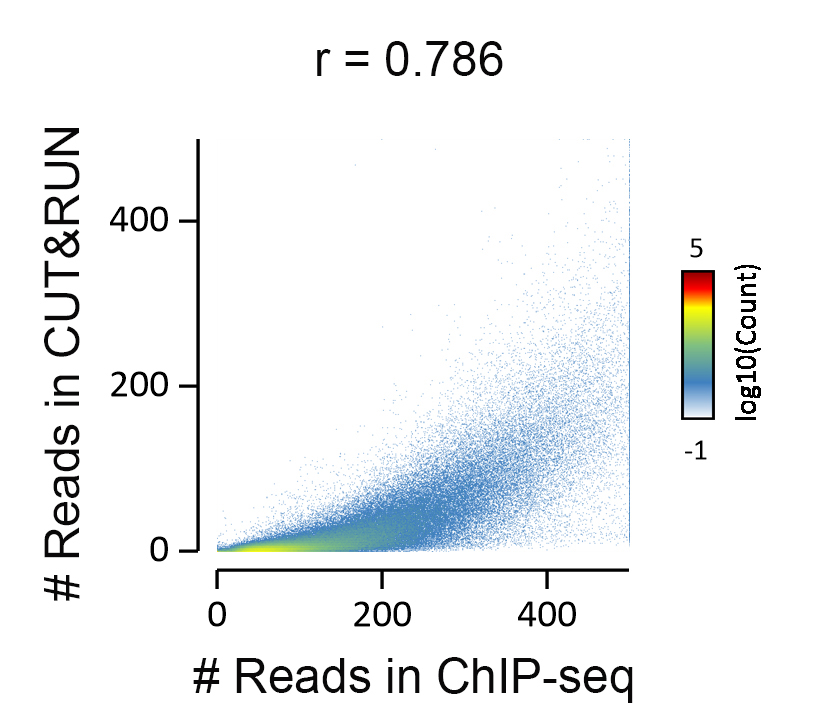
{"url":"https://www.epicypher.com/products/nucleosomes/snap-chip-sup-sup-spike-ins/snap-chip-sup-sup-certified-antibodies/histone-h3k36me3-antibody-snap-chip-certified-cutana-cut-run-compatible-discontinued","add_this":[{"service":"facebook","annotation":""},{"service":"email","annotation":""},{"service":"print","annotation":""},{"service":"twitter","annotation":""},{"service":"linkedin","annotation":""}],"warranty":"","gtin":"","max_purchase_quantity":0,"id":"629","can_purchase":false,"meta_description":"Histone H3K36me3 Antibody, SNAP-ChIP® Certified, CUTANA™ CUT&RUN Compatible (13-0031)","category":[],"meta_keywords":"Histone H3K36me3 Antibody - SNAP-ChIP® Certified, 13-0031, H3K36me3, SNAP, H3K36me3 SNAP ChIP, SNAP ChIP, H3K36me3 SNAP-ChIP, SNAP-ChIP, H3K36me3 SNAP, H3K36me3 Antibody, CUTANA™ compatible, CUT&RUN, cut and run, cleavage under targets and release using nuclease, 13-0031","AddThisServiceButtonMeta":"","images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/629/581/Screen_Shot_2018-09-25_at_2.00.27_PM__10558.1538135192.png?c=2","alt":"Histone H3K36me3 Antibody, SNAP-ChIP® Certified, CUTANA™ CUT&RUN Compatible *DISCONTINUED*"}],"main_image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/629/581/Screen_Shot_2018-09-25_at_2.00.27_PM__10558.1538135192.png?c=2","alt":"Histone H3K36me3 Antibody, SNAP-ChIP® Certified, CUTANA™ CUT&RUN Compatible *DISCONTINUED*"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=629","shipping":[],"num_reviews":0,"weight":"0.00 LBS","custom_fields":[{"id":"446","name":"Pack Size","value":"100 μg"}],"sku":"13-0031","description":"<table cellpadding=\"2\" cellspacing=\"2\" style=\"width: 100%\">\n <a\n style=\"color: #fff\"\n href=\"/products/antibodies/cut-and-run-antibodies/cut-and-run-antibodies-histone-ptms/h3k36me3-antibody-snap-certified-for-cut-and-run\">\n <p\n style=\"\n background-color: #4698cb;\n color: #fff;\n padding: 1.3rem;\n text-align: center;\n border-radius: 12px;\n margin-top: 2.5rem;\n \">\n Product Discontinued - see 13-0058 as a recommended alternative\n </p>\n <tbody>\n <tr valign=\"top\">\n <td>\n <table cellpadding=\"1\" cellspacing=\"1\" style=\"width: 95%\">\n <tbody>\n <tr>\n <td valign=\"top\">\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><strong\n >Histone H3K36me3 Antibody: SNAP-ChIP Certified, CUTANA™\n Compatible Description:</strong\n >\n This antibody meets EpiCypher’s “SNAP-ChIP®\n Certified” criteria for specificity and target\n enrichment in ChIP (<20% cross-reactivity to related\n histone post-translation modifications and >5% recovery of\n target input determined using SNAP-ChIP K-MetStat Panel\n spike-in controls; EpiCypher Catalog No. 19-1001). Although\n its specificity in CUT&RUN has yet to be empirically\n determined <i>in situ</i> using spike-in controls, CUT&RUN\n data produced by this antibody shows a genome-wide enrichment\n pattern characteristic of H3K36me3 and is highly correlated\n with ChIP-seq (Figures 3-5). <br /></span\n ><br />\n </td>\n </tr>\n <tr>\n <td valign=\"top\">\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><br /><strong\n >Histone H3K36me3 Antibody, SNAP-ChIP Certified, CUTANA™\n Compatible Certified Immunogen:</strong\n >\n A synthetic peptide corresponding to histone H3 trimethylated\n at lysine 36.</span\n >\n <p></p>\n </td>\n </tr>\n <tr>\n <td valign=\"top\">\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><strong\n >Histone H3K36me3 Antibody, SNAP-ChIP Certified, CUTANA™\n Compatible Certified Formulation:</strong\n >\n Protein A affinity-purified antibody in PBS, pH 7.2 with 0.09%\n sodium azide, 1% BSA, and 50% glycerol.</span\n >\n <p></p>\n </td>\n </tr>\n <tr>\n <td valign=\"top\">\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><strong\n >Histone H3K36me3 Antibody, SNAP-ChIP Certified, CUTANA™\n Compatible Certified Storage and Stability:</strong\n >\n Stable for 1 years at -20°C from date of receipt.</span\n >\n <p></p>\n </td>\n </tr>\n\n <tr>\n <td valign=\"top\">\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><strong\n >Histone H3K36me3 Antibody, SNAP-ChIP Certified, CUTANA™\n Compatible Certified Application Note:<br />\n Recommended Diltutions:\n </strong>\n <strong\n ><br />\n ChIP/ChIP-seq:</strong\n >\n 2 - 5 μg per 5 µg chromatin\n <strong\n ><br />\n CUT&RUN:</strong\n >\n 1:100\n <strong\n ><br />\n L:</strong\n >\n 1:4000 dilution<br /><strong>WB:</strong> 0.5 - 2 µg/mL</span\n >\n <p></p>\n </td>\n </tr>\n\n <tr>\n <td valign=\"top\">\n <p>\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><span style=\"font-size: 12pt\"\n ><strong\n >Histone H3K36me3 Antibody, SNAP-ChIP Certified, CUTANA™\n Compatible References:</strong\n ></span\n ></span\n >\n </p>\n\n <p></p>\n\n <p>\n <span style=\"font-family: arial, helvetica, sans-serif\"\n >Grzybowski et al (2015) Mol Cell 58:886</span\n >\n </p>\n\n <p>\n <span style=\"font-family: arial, helvetica, sans-serif\"\n >Shah et al (2018) Mol Cell 72:162</span\n >\n </p>\n </td>\n </tr>\n <tr>\n <td valign=\"top\">\n <span style=\"font-family: arial, helvetica, sans-serif\"\n >View technical datasheet for this product.\n <a\n href=\"https://www.epicypher.com/content/documents/tds/13-0031.pdf\"\n target=\"_new\">\n <img\n alt=\"13-0031 Datasheet\"\n height=\"40\"\n src=\"https://cdn7.bigcommerce.com/s-y9o92/content/documents/tds/icon.png\"\n width=\"30\" /></a\n ></span>\n </td>\n </tr>\n <tr>\n <td>\n <p></p>\n\n <p>\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><span style=\"font-size: 10pt\"\n ><strong>Applications Key: ChIP</strong>-Chromatin\n IP; <strong>E</strong>-ELISA; <strong>FACS</strong>-Flow\n cytometry; <strong>IF</strong>-Immunofluorescence;\n <strong>IHC</strong>-Immunohistochemistry;\n <strong>ICC</strong>-Immunocytochemistry;\n <strong>L</strong>-Luminex;\n <strong>IP</strong>-Immunoprecipitation;\n <strong>WB</strong>-Western Blotting</span\n ></span\n >\n </p>\n\n <p>\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><span style=\"font-size: 10pt\"\n ><strong>Reactivity Key: </strong\n ><strong>B</strong>-Bovine; <strong>Ce</strong>-C.\n elegans; <strong>Ch</strong>-Chicken; <strong>Dm</strong>-\n Drosophila; <strong>Eu</strong>-Eukaryote;\n <strong>H</strong>-Human; <strong>M</strong>-Mouse;\n <strong>Ma</strong>-Mammal; <strong>R</strong>-Rat;\n <strong>Sc</strong>-S.cerevesiae; <strong>Sp</strong>-S.\n pombe; <strong>WR</strong>-Wide Range (predicted);\n <strong>X</strong>-Xenopus;\n <strong>Z</strong>-Zebrafish</span\n ></span\n >\n </p>\n </td>\n </tr>\n </tbody>\n </table>\n </td>\n <td>\n <table cellpadding=\"3\" cellspacing=\"3\" style=\"width: 300px\">\n <tbody>\n <tr>\n <td>\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><a\n href=\"https://cdn11.bigcommerce.com/s-y9o92/product_images/uploaded_images/h3k36me3-epicypher-13-0031.jpg?t=1600974099&_ga=2.224614169.241922123.1600715432-402741698.1598987696\"\n target=\"_new\"\n ><img\n alt=\"13-0031 Representative SNAP-ChIP-seq results: \"\n height=\"100\"\n src=\"https://cdn11.bigcommerce.com/s-y9o92/product_images/uploaded_images/h3k36me3-epicypher-13-0031.jpg?t=1600974099&_ga=2.224614169.241922123.1600715432-402741698.1598987696\"\n width=\"150\" /></a\n ></span>\n </td>\n </tr>\n <tr>\n <td valign=\"top\">\n <p>\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><strong>Representative SNAP-ChIP-seq results: </strong\n >Cumulative histogram plot and heatmap of signal intensity\n depict H3K36me3 ChIP-seq data aligned to gene bodies (gene\n start - end, +/- 3.0 kb; left). Two representative genomic\n regions depicting H3K36me3 peak structure and distribution\n are shown (right). Data shown are representative of H3K36me3\n ChIP antibody (EpiCypher Catalog No. 13-0031) and are not\n lot-specific. Native ChIP-seq was performed using K562 cells\n as described (<a\n href=\"https://pubmed.ncbi.nlm.nih.gov/30244833/\"\n target=\"_blank\"\n >Shah et al., Mol Cell 2018</a\n >) with SNAP-ChIP<sup>TM</sup> K-MetStat Spike-in (Catalog\n No.\n <a\n href=\"https://www.epicypher.com/products/nucleosomes/snap-chip-k-metstat-panel\"\n target=\"_blank\"\n >19-1001</a\n >) nucleosome controls added prior to chromatin digestion to\n confirm antibody specificity and ChIP efficiency. Paired-end\n sequencing libraries were prepared using the NEBNext<sup\n >®</sup\n >\n Ultra<sup>TM</sup> II DNA Library Prep Kit for\n Illumina<sup>®</sup>. ChIP libraries were sequenced on an\n Illumina<sup>®</sup> NextSeq. Sequencing reads were aligned\n to the human genome using Bowtie 2 (Johns Hopkins\n University). Bigwig files of read enrichment in binned\n genomic regions (signal intensity) flanking the indicated\n gene features were used to create a cumulative histogram\n plot and heatmap of signal intensity (<a\n href=\"https://basepairtech.com/\"\n target=\"_blank\">\n www.basepairtech.com</a\n >). Gene browser shots were generated using the Integrative\n Genomics Viewer (IGV, Broad Institute) with the window size\n denoted (top). <br />\n <strong>(Click image to enlarge) </strong></span\n >\n </p>\n\n <p></p>\n </td>\n </tr>\n <tr>\n <td>\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><a\n href=\"https://www.epicypher.com/content/images/products/antibodies/13-0031_luminex.jpg\"\n target=\"_new\"\n ><img\n alt=\"13-0031 Luminex Data\"\n height=\"100\"\n src=\"https://cdn11.bigcommerce.com/s-y9o92/content/images/products/antibodies/13-0031_luminex.jpg\"\n width=\"150\" /></a\n ></span>\n </td>\n </tr>\n <tr>\n <td valign=\"top\">\n <p>\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><strong>Luminex Mutliplexed Specificity Profiling: </strong\n >Histone H3K36me3 antibody was assessed using a Luminex®\n based approach employing dCypher® Nucleosome K-MetStat\n Panel (EpiCypher Catalog No. 16-9002). The panel comprises\n biotinylated designer nucleosomes (x-axis) individually\n coupled to color coded Luminex Magplex® beads. Antibody\n binding to the panel of 16 nucleosomes was tested in\n multiplex at a 1:1000 dilution, and detected with second\n layer anti-IgG*PE. Data was generated using a Luminex\n FlexMAP3D®. Data normalized to relevant on-target\n (H3K36me3; set to 100) <br />\n <strong>(Click image to enlarge) </strong></span\n >\n </p>\n\n <p></p>\n </td>\n </tr>\n <!--BEGIN IMAGE AND CAPTION SECTION-->\n <tr>\n <td>\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><a\n href=\"https://www.epicypher.com/content/images/products/antibodies/13-0031_SNAP_qPCR.jpg\"\n target=\"_new\"\n ><img\n alt=\"13-0031 SNAP-ChIP qPCR Data\"\n height=\"100\"\n src=\"https://cdn11.bigcommerce.com/s-y9o92/content/images/products/antibodies/13-0031_SNAP_qPCR.jpg\"\n width=\"200\" /></a\n ></span>\n </td>\n </tr>\n <tr>\n <td valign=\"top\">\n <p>\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><strong>SNAP-ChIP Data:</strong> Histone H3K36me3 antibody\n (5 µg) was tested in a native ChIP experiment using\n chromatin from K-562 cells (5 µg) with the SNAP-ChIP\n K-MetStat Panel (EpiCypher Catalog No. 19-1001) spiked-in\n prior to micrococcal nuclease digestion. Specificity (left\n y-axis) was determined by qPCR for the DNA barcodes\n corresponding to modified nucleosomes in the SNAP-ChIP panel\n (x-axis). Black bar represents antibody efficiency (right\n y-axis; log scale) and indicates percentage of the target\n immunoprecipitated relative to input. Error bars represent\n mean ± SEM in replicate ChIP experiments. <br />\n <strong>(Click image to enlarge) </strong></span\n >\n </p>\n\n <p></p>\n </td>\n </tr>\n <!--END IMAGE AND CAPTION SECTION-->\n <tr valign=\"top\">\n <td valign=\"top\">\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><a\n href=\"https://www.epicypher.com/content/images/products/antibodies/13-0031_seq_tracks.jpg\"\n target=\"_new\"\n ><img\n alt=\"13-0031 Representative Sequencing Tracks\"\n height=\"100\"\n src=\"https://cdn11.bigcommerce.com/s-y9o92/content/images/products/antibodies/13-0031_seq_tracks.jpg\"\n width=\"350\" /></a\n ></span>\n </td>\n </tr>\n <tr>\n <td valign=\"top\">\n <p>\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><strong\n >H3K36me3 SNAP-ChIP-seq and CUT&RUN representative tracks: </strong\n >A gene browser shot generated using the Integrative\n Genomics Viewer (IGV, Broad Institute) shows a\n representative locus for EpiCypher H3K36me3 ChIP-seq (blue\n tracks, 3 μg antibody) and CUT&RUN (green track, 1:100\n antibody dilution). For comparison ENCODE H3K36me3 ChIP-seq\n using a different antibody is shown (bottom orange track,\n GEO accession number GSM621387). Similar results in peak\n structure and location were observed throughout the genome\n for EpiCypher H3K36me3 antibody in ChIP-seq and CUT&RUN.\n <i\n >Methods: Native ChIP-seq was performed as described (Shah\n et al., Mol Cell 2018). CUT&RUN was performed using\n EpiCypher CUTANA pAG-MNase for ChIC/CUT&RUN (EpiCypher\n Catalog No. 15-1016) as described\n (EpiCypher.com/cutana-protocol). Library preparation was\n performed with 10 ng DNA using the NEBNext<sup>®</sup>\n Ultra<sup>TM</sup> II DNA Library Prep Kit for\n Illumina<sup>®</sup>. ChIP libraries were sequenced on an\n Illumina NextSeq 550 (2x150bp paired end). The total\n number of reads was 33.6 million for ChIP-seq and 3.2\n million for CUT&RUN.</i\n >\n <br />\n <strong>(Click image to enlarge) </strong></span\n >\n </p>\n\n <p></p>\n </td>\n </tr>\n <tr valign=\"top\">\n <td valign=\"top\">\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><a\n href=\"https://www.epicypher.com/content/images/products/antibodies/13-0031_GWA.jpg\"\n target=\"_new\"\n ><img\n alt=\"13-0031 Genome Wide Analysis\"\n height=\"100\"\n src=\"https://cdn11.bigcommerce.com/s-y9o92/content/images/products/antibodies/13-0031_GWA.jpg\"\n width=\"127\" /></a\n ></span>\n </td>\n </tr>\n <tr>\n <td valign=\"top\">\n <p>\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><strong>ChIP-seq and CUT&RUN genome wide analysis </strong>\n EpiCypher H3K36me3 antibody was tested in native ChIP-seq\n <strong>(A)</strong> and CUT&RUN <strong>(B)</strong> using\n the methods described above. Genome-wide analysis of\n H3K36me3 enrichment (signal intensity) flanking annotated\n genes (gene start to gene end; +/- 3kb) is graphed as a\n cumulative histogram plot (top) and shown in a heatmap\n (bottom). Individual gene loci in each row of the heatmap\n are colored by signal intensity and sorted by strongest to\n lowest enrichment (top to bottom). EpiCypher H3K36me3\n antibody displays a characteristic enrichment pattern in\n gene bodies.<br />\n <strong>(Click image to enlarge) </strong></span\n >\n </p>\n\n <p></p>\n </td>\n </tr>\n <tr valign=\"top\">\n <td valign=\"top\">\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><a\n href=\"https://www.epicypher.com/content/images/products/antibodies/13-0031_correlation.jpg\"\n target=\"_new\"\n ><img\n alt=\"13-0031 Correlation Analysis\"\n height=\"100\"\n src=\"https://cdn11.bigcommerce.com/s-y9o92/content/images/products/antibodies/13-0031_correlation.jpg\"\n width=\"127\" /></a\n ></span>\n </td>\n </tr>\n <tr>\n <td valign=\"top\">\n <p>\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><strong>ChIP-seq vs. CUT&RUN correlation analysis </strong>\n Genome-wide correlation analysis was performed to compare\n EpiCypher H3K36me3 antibody enrichment in ChIP-seq and\n CUT&RUN. The number of reads per 5 kb binned region across\n the genome is plotted for CUT&RUN (x-axis) vs. ChIP-seq\n (y-axis) (EaSeq). ChIP-seq and CUT&RUN data generated using\n this antibody are highly correlated (Pearson correlation r =\n 0.786).<br />\n <strong>(Click image to enlarge) </strong></span\n >\n </p>\n\n <p></p>\n </td>\n </tr>\n <tr valign=\"top\">\n <td valign=\"top\">\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><a\n href=\"https://www.epicypher.com/content/images/products/antibodies/13-0031_wb.jpg\"\n target=\"_new\"\n ><img\n alt=\"13-0031 Western Blot\"\n height=\"100\"\n src=\"https://cdn11.bigcommerce.com/s-y9o92/content/images/products/antibodies/13-0031_wb.jpg\"\n width=\"127\" /></a\n ></span>\n </td>\n </tr>\n <tr>\n <td valign=\"top\">\n <p>\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><strong>Western blot analysis </strong> Recombinant histone\n H3.3 (Lane 1) and acid extracts of HeLa cells (Lane 2) were\n blotted onto PVDF and probed with 2 μg/mL Histone H3K36me3\n antibody.<br />\n <strong>(Click image to enlarge) </strong></span\n >\n </p>\n\n <p></p>\n </td>\n </tr>\n </tbody>\n </table>\n </td>\n </tr>\n </tbody>\n</table>\n<style>\n .button-group {\n display: none;\n }\n</style>","tags":[],"detail_messages":"","availability":"","page_title":"Histone H3K36me3 Antibody, SNAP-ChIP® Certified, CUTANA™ CUT&RUN Compatible (13-0031)","mpn":"","upc":null,"options":[],"related_products":[{"id":411,"sku":null,"name":"SNAP-ChIP® K-MetStat Panel","url":"https://www.epicypher.com/products/nucleosomes/snap-chip-k-metstat-panel","availability":"","rating":null,"brand":{"name":null},"category":["Nucleosomes","Nucleosomes/SNAP-ChIP<sup>®</sup> Spike-ins"],"summary":"\n\t\n\t\t\n\t\t\t\n\t\t\t\n\t\t\t\t\n\t\t\t\t\t\n\t\t\t\t\t\t\n\t\t\t\t\t\t\n\n\t\t\t\t\t\t\n\t\t\t\t\t\tSNAP-ChIP K-MetStat Panel Description: A panel ","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/411/282/SNAP_Chip__53961.1569012494.png?c=2","alt":"SNAP-ChIP® K-MetStat Panel"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/411/282/SNAP_Chip__53961.1569012494.png?c=2","alt":"SNAP-ChIP® K-MetStat Panel"}],"date_added":"18th Sep 2017","pre_order":false,"show_cart_action":true,"has_options":true,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":null,"num_reviews":null,"weight":{"formatted":"0.01 LBS","value":0.01},"demo":false,"price":{"without_tax":{"currency":"USD","formatted":"$365.00","value":365},"price_range":{"min":{"without_tax":{"currency":"USD","formatted":"$365.00","value":365},"tax_label":"Sales Tax"},"max":{"without_tax":{"currency":"USD","formatted":"$2,795.00","value":2795},"tax_label":"Sales Tax"}},"tax_label":"Sales Tax"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=411"},{"id":555,"sku":null,"name":"SNAP-ChIP® OncoStat Panel","url":"https://www.epicypher.com/products/nucleosomes/snap-chip-oncostat-panel","availability":"","rating":null,"brand":{"name":null},"category":["Nucleosomes","Nucleosomes/SNAP-ChIP<sup>®</sup> Spike-ins"],"summary":" \n\n ","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/555/399/OncoStat__15594.1569012525.png?c=2","alt":"SNAP-ChIP® OncoStat Panel"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/555/399/OncoStat__15594.1569012525.png?c=2","alt":"SNAP-ChIP® OncoStat Panel"}],"date_added":"31st Jul 2018","pre_order":false,"show_cart_action":true,"has_options":true,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":null,"num_reviews":null,"weight":{"formatted":"0.01 LBS","value":0.01},"demo":false,"price":{"without_tax":{"currency":"USD","formatted":"$365.00","value":365},"price_range":{"min":{"without_tax":{"currency":"USD","formatted":"$365.00","value":365},"tax_label":"Sales Tax"},"max":{"without_tax":{"currency":"USD","formatted":"$2,795.00","value":2795},"tax_label":"Sales Tax"}},"tax_label":"Sales Tax"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=555"},{"id":626,"sku":"13-0028","name":"Histone H3K4me3 Antibody, SNAP-ChIP Certified *DISCONTINUED*","url":"https://www.epicypher.com/products/antibodies/snap-chip-certified-antibodies/histone-h3k4me3-antibody-snap-chip-certified","availability":"","rating":null,"brand":{"name":null},"category":[],"summary":"\n \n \n \n ","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/626/578/Screen_Shot_2018-09-25_at_2.00.27_PM__52796.1538135192.png?c=2","alt":"Histone H3K4me3 Antibody, SNAP-ChIP Certified *DISCONTINUED*"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/626/578/Screen_Shot_2018-09-25_at_2.00.27_PM__52796.1538135192.png?c=2","alt":"Histone H3K4me3 Antibody, SNAP-ChIP Certified *DISCONTINUED*"}],"date_added":"24th Sep 2018","pre_order":false,"show_cart_action":false,"has_options":false,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":[{"id":452,"name":"Pack Size","value":"100 μg"}],"num_reviews":null,"weight":{"formatted":"0.00 LBS","value":0},"demo":false,"add_to_wishlist_url":"/wishlist.php?action=add&product_id=626"},{"id":625,"sku":"13-0027","name":"Histone H3K4me2 Antibody, SNAP-Certified™ for CUT&RUN","url":"https://www.epicypher.com/products/antibodies/cutana-cut-run-antibodies/cut-run-antibodies-histone-ptms/histone-h3k4me2-antibody-snap-certified-for-cut-run","availability":"","rating":null,"brand":{"name":null},"category":["Antibodies/CUTANA™ CUT&RUN Antibodies","Antibodies/CUTANA™ CUT&RUN Antibodies/CUTANA™ CUT&RUN Antibodies to Histone PTMs","Epigenetics Kits and Reagents/CUTANA™ ChIC / CUT&RUN Assays"],"summary":"\n \n \n Type: Monoclonal\n \n \n Target Size: 15 kDa\n \n \n Format: Affi","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/625/577/Screen_Shot_2018-09-25_at_2.00.27_PM__04625.1538135192.png?c=2","alt":"Histone H3K4me2 Antibody, SNAP-Certified™ for CUT&RUN"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/625/577/Screen_Shot_2018-09-25_at_2.00.27_PM__04625.1538135192.png?c=2","alt":"Histone H3K4me2 Antibody, SNAP-Certified™ for CUT&RUN"}],"date_added":"24th Sep 2018","pre_order":false,"show_cart_action":true,"has_options":false,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":[{"id":451,"name":"Pack Size","value":"100 μg"}],"num_reviews":null,"weight":{"formatted":"0.00 LBS","value":0},"demo":false,"add_to_cart_url":"https://www.epicypher.com/cart.php?action=add&product_id=625","price":{"without_tax":{"currency":"USD","formatted":"$525.00","value":525},"tax_label":"Sales Tax"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=625"},{"id":628,"sku":"13-0030","name":"Histone H3K27me3 Antibody, SNAP-ChIP® Certified, CUTANA™ CUT&RUN Compatible *DISCONTINUED*","url":"https://www.epicypher.com/products/antibodies/snap-chip-certified-antibodies/histone-h3k27me3-antibody-snap-chip-certified-cutana-cut-run-compatible-discontinued","availability":"","rating":null,"brand":{"name":null},"category":[],"summary":"\n \n ","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/628/940/histone-h3k27me3-antibody-snap-chip-certified-cutana-cutandrun-compatible-discontinued__17318.1645734531.jpg?c=2","alt":"Histone H3K27me3 Antibody, SNAP-ChIP Certified, CUTANA CUTandRUN Compatible - DISCONTINUED"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/628/940/histone-h3k27me3-antibody-snap-chip-certified-cutana-cutandrun-compatible-discontinued__17318.1645734531.jpg?c=2","alt":"Histone H3K27me3 Antibody, SNAP-ChIP Certified, CUTANA CUTandRUN Compatible - DISCONTINUED"}],"date_added":"24th Sep 2018","pre_order":false,"show_cart_action":false,"has_options":false,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":[{"id":470,"name":"Pack Size","value":"100 μg"}],"num_reviews":null,"weight":{"formatted":"0.00 LBS","value":0},"demo":false,"add_to_wishlist_url":"/wishlist.php?action=add&product_id=628"}],"shipping_messages":[],"rating":0,"title":"Histone H3K36me3 Antibody, SNAP-ChIP® Certified, CUTANA™ CUT&RUN Compatible *DISCONTINUED*","gift_wrapping_available":false,"min_purchase_quantity":0} Pack Size: 100 μg
{"url":"https://www.epicypher.com/products/nucleosomes/snap-chip-sup-sup-spike-ins/snap-chip-sup-sup-certified-antibodies/histone-h3k36me3-antibody-snap-chip-certified-cutana-cut-run-compatible-discontinued","add_this":[{"service":"facebook","annotation":""},{"service":"email","annotation":""},{"service":"print","annotation":""},{"service":"twitter","annotation":""},{"service":"linkedin","annotation":""}],"warranty":"","gtin":"","max_purchase_quantity":0,"id":"629","can_purchase":false,"meta_description":"Histone H3K36me3 Antibody, SNAP-ChIP® Certified, CUTANA™ CUT&RUN Compatible (13-0031)","category":[],"meta_keywords":"Histone H3K36me3 Antibody - SNAP-ChIP® Certified, 13-0031, H3K36me3, SNAP, H3K36me3 SNAP ChIP, SNAP ChIP, H3K36me3 SNAP-ChIP, SNAP-ChIP, H3K36me3 SNAP, H3K36me3 Antibody, CUTANA™ compatible, CUT&RUN, cut and run, cleavage under targets and release using nuclease, 13-0031","AddThisServiceButtonMeta":"","images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/629/581/Screen_Shot_2018-09-25_at_2.00.27_PM__10558.1538135192.png?c=2","alt":"Histone H3K36me3 Antibody, SNAP-ChIP® Certified, CUTANA™ CUT&RUN Compatible *DISCONTINUED*"}],"main_image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/629/581/Screen_Shot_2018-09-25_at_2.00.27_PM__10558.1538135192.png?c=2","alt":"Histone H3K36me3 Antibody, SNAP-ChIP® Certified, CUTANA™ CUT&RUN Compatible *DISCONTINUED*"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=629","shipping":[],"num_reviews":0,"weight":"0.00 LBS","custom_fields":[{"id":"446","name":"Pack Size","value":"100 μg"}],"sku":"13-0031","description":"<table cellpadding=\"2\" cellspacing=\"2\" style=\"width: 100%\">\n <a\n style=\"color: #fff\"\n href=\"/products/antibodies/cut-and-run-antibodies/cut-and-run-antibodies-histone-ptms/h3k36me3-antibody-snap-certified-for-cut-and-run\">\n <p\n style=\"\n background-color: #4698cb;\n color: #fff;\n padding: 1.3rem;\n text-align: center;\n border-radius: 12px;\n margin-top: 2.5rem;\n \">\n Product Discontinued - see 13-0058 as a recommended alternative\n </p>\n <tbody>\n <tr valign=\"top\">\n <td>\n <table cellpadding=\"1\" cellspacing=\"1\" style=\"width: 95%\">\n <tbody>\n <tr>\n <td valign=\"top\">\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><strong\n >Histone H3K36me3 Antibody: SNAP-ChIP Certified, CUTANA™\n Compatible Description:</strong\n >\n This antibody meets EpiCypher’s “SNAP-ChIP®\n Certified” criteria for specificity and target\n enrichment in ChIP (<20% cross-reactivity to related\n histone post-translation modifications and >5% recovery of\n target input determined using SNAP-ChIP K-MetStat Panel\n spike-in controls; EpiCypher Catalog No. 19-1001). Although\n its specificity in CUT&RUN has yet to be empirically\n determined <i>in situ</i> using spike-in controls, CUT&RUN\n data produced by this antibody shows a genome-wide enrichment\n pattern characteristic of H3K36me3 and is highly correlated\n with ChIP-seq (Figures 3-5). <br /></span\n ><br />\n </td>\n </tr>\n <tr>\n <td valign=\"top\">\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><br /><strong\n >Histone H3K36me3 Antibody, SNAP-ChIP Certified, CUTANA™\n Compatible Certified Immunogen:</strong\n >\n A synthetic peptide corresponding to histone H3 trimethylated\n at lysine 36.</span\n >\n <p></p>\n </td>\n </tr>\n <tr>\n <td valign=\"top\">\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><strong\n >Histone H3K36me3 Antibody, SNAP-ChIP Certified, CUTANA™\n Compatible Certified Formulation:</strong\n >\n Protein A affinity-purified antibody in PBS, pH 7.2 with 0.09%\n sodium azide, 1% BSA, and 50% glycerol.</span\n >\n <p></p>\n </td>\n </tr>\n <tr>\n <td valign=\"top\">\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><strong\n >Histone H3K36me3 Antibody, SNAP-ChIP Certified, CUTANA™\n Compatible Certified Storage and Stability:</strong\n >\n Stable for 1 years at -20°C from date of receipt.</span\n >\n <p></p>\n </td>\n </tr>\n\n <tr>\n <td valign=\"top\">\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><strong\n >Histone H3K36me3 Antibody, SNAP-ChIP Certified, CUTANA™\n Compatible Certified Application Note:<br />\n Recommended Diltutions:\n </strong>\n <strong\n ><br />\n ChIP/ChIP-seq:</strong\n >\n 2 - 5 μg per 5 µg chromatin\n <strong\n ><br />\n CUT&RUN:</strong\n >\n 1:100\n <strong\n ><br />\n L:</strong\n >\n 1:4000 dilution<br /><strong>WB:</strong> 0.5 - 2 µg/mL</span\n >\n <p></p>\n </td>\n </tr>\n\n <tr>\n <td valign=\"top\">\n <p>\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><span style=\"font-size: 12pt\"\n ><strong\n >Histone H3K36me3 Antibody, SNAP-ChIP Certified, CUTANA™\n Compatible References:</strong\n ></span\n ></span\n >\n </p>\n\n <p></p>\n\n <p>\n <span style=\"font-family: arial, helvetica, sans-serif\"\n >Grzybowski et al (2015) Mol Cell 58:886</span\n >\n </p>\n\n <p>\n <span style=\"font-family: arial, helvetica, sans-serif\"\n >Shah et al (2018) Mol Cell 72:162</span\n >\n </p>\n </td>\n </tr>\n <tr>\n <td valign=\"top\">\n <span style=\"font-family: arial, helvetica, sans-serif\"\n >View technical datasheet for this product.\n <a\n href=\"https://www.epicypher.com/content/documents/tds/13-0031.pdf\"\n target=\"_new\">\n <img\n alt=\"13-0031 Datasheet\"\n height=\"40\"\n src=\"https://cdn7.bigcommerce.com/s-y9o92/content/documents/tds/icon.png\"\n width=\"30\" /></a\n ></span>\n </td>\n </tr>\n <tr>\n <td>\n <p></p>\n\n <p>\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><span style=\"font-size: 10pt\"\n ><strong>Applications Key: ChIP</strong>-Chromatin\n IP; <strong>E</strong>-ELISA; <strong>FACS</strong>-Flow\n cytometry; <strong>IF</strong>-Immunofluorescence;\n <strong>IHC</strong>-Immunohistochemistry;\n <strong>ICC</strong>-Immunocytochemistry;\n <strong>L</strong>-Luminex;\n <strong>IP</strong>-Immunoprecipitation;\n <strong>WB</strong>-Western Blotting</span\n ></span\n >\n </p>\n\n <p>\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><span style=\"font-size: 10pt\"\n ><strong>Reactivity Key: </strong\n ><strong>B</strong>-Bovine; <strong>Ce</strong>-C.\n elegans; <strong>Ch</strong>-Chicken; <strong>Dm</strong>-\n Drosophila; <strong>Eu</strong>-Eukaryote;\n <strong>H</strong>-Human; <strong>M</strong>-Mouse;\n <strong>Ma</strong>-Mammal; <strong>R</strong>-Rat;\n <strong>Sc</strong>-S.cerevesiae; <strong>Sp</strong>-S.\n pombe; <strong>WR</strong>-Wide Range (predicted);\n <strong>X</strong>-Xenopus;\n <strong>Z</strong>-Zebrafish</span\n ></span\n >\n </p>\n </td>\n </tr>\n </tbody>\n </table>\n </td>\n <td>\n <table cellpadding=\"3\" cellspacing=\"3\" style=\"width: 300px\">\n <tbody>\n <tr>\n <td>\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><a\n href=\"https://cdn11.bigcommerce.com/s-y9o92/product_images/uploaded_images/h3k36me3-epicypher-13-0031.jpg?t=1600974099&_ga=2.224614169.241922123.1600715432-402741698.1598987696\"\n target=\"_new\"\n ><img\n alt=\"13-0031 Representative SNAP-ChIP-seq results: \"\n height=\"100\"\n src=\"https://cdn11.bigcommerce.com/s-y9o92/product_images/uploaded_images/h3k36me3-epicypher-13-0031.jpg?t=1600974099&_ga=2.224614169.241922123.1600715432-402741698.1598987696\"\n width=\"150\" /></a\n ></span>\n </td>\n </tr>\n <tr>\n <td valign=\"top\">\n <p>\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><strong>Representative SNAP-ChIP-seq results: </strong\n >Cumulative histogram plot and heatmap of signal intensity\n depict H3K36me3 ChIP-seq data aligned to gene bodies (gene\n start - end, +/- 3.0 kb; left). Two representative genomic\n regions depicting H3K36me3 peak structure and distribution\n are shown (right). Data shown are representative of H3K36me3\n ChIP antibody (EpiCypher Catalog No. 13-0031) and are not\n lot-specific. Native ChIP-seq was performed using K562 cells\n as described (<a\n href=\"https://pubmed.ncbi.nlm.nih.gov/30244833/\"\n target=\"_blank\"\n >Shah et al., Mol Cell 2018</a\n >) with SNAP-ChIP<sup>TM</sup> K-MetStat Spike-in (Catalog\n No.\n <a\n href=\"https://www.epicypher.com/products/nucleosomes/snap-chip-k-metstat-panel\"\n target=\"_blank\"\n >19-1001</a\n >) nucleosome controls added prior to chromatin digestion to\n confirm antibody specificity and ChIP efficiency. Paired-end\n sequencing libraries were prepared using the NEBNext<sup\n >®</sup\n >\n Ultra<sup>TM</sup> II DNA Library Prep Kit for\n Illumina<sup>®</sup>. ChIP libraries were sequenced on an\n Illumina<sup>®</sup> NextSeq. Sequencing reads were aligned\n to the human genome using Bowtie 2 (Johns Hopkins\n University). Bigwig files of read enrichment in binned\n genomic regions (signal intensity) flanking the indicated\n gene features were used to create a cumulative histogram\n plot and heatmap of signal intensity (<a\n href=\"https://basepairtech.com/\"\n target=\"_blank\">\n www.basepairtech.com</a\n >). Gene browser shots were generated using the Integrative\n Genomics Viewer (IGV, Broad Institute) with the window size\n denoted (top). <br />\n <strong>(Click image to enlarge) </strong></span\n >\n </p>\n\n <p></p>\n </td>\n </tr>\n <tr>\n <td>\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><a\n href=\"https://www.epicypher.com/content/images/products/antibodies/13-0031_luminex.jpg\"\n target=\"_new\"\n ><img\n alt=\"13-0031 Luminex Data\"\n height=\"100\"\n src=\"https://cdn11.bigcommerce.com/s-y9o92/content/images/products/antibodies/13-0031_luminex.jpg\"\n width=\"150\" /></a\n ></span>\n </td>\n </tr>\n <tr>\n <td valign=\"top\">\n <p>\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><strong>Luminex Mutliplexed Specificity Profiling: </strong\n >Histone H3K36me3 antibody was assessed using a Luminex®\n based approach employing dCypher® Nucleosome K-MetStat\n Panel (EpiCypher Catalog No. 16-9002). The panel comprises\n biotinylated designer nucleosomes (x-axis) individually\n coupled to color coded Luminex Magplex® beads. Antibody\n binding to the panel of 16 nucleosomes was tested in\n multiplex at a 1:1000 dilution, and detected with second\n layer anti-IgG*PE. Data was generated using a Luminex\n FlexMAP3D®. Data normalized to relevant on-target\n (H3K36me3; set to 100) <br />\n <strong>(Click image to enlarge) </strong></span\n >\n </p>\n\n <p></p>\n </td>\n </tr>\n <!--BEGIN IMAGE AND CAPTION SECTION-->\n <tr>\n <td>\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><a\n href=\"https://www.epicypher.com/content/images/products/antibodies/13-0031_SNAP_qPCR.jpg\"\n target=\"_new\"\n ><img\n alt=\"13-0031 SNAP-ChIP qPCR Data\"\n height=\"100\"\n src=\"https://cdn11.bigcommerce.com/s-y9o92/content/images/products/antibodies/13-0031_SNAP_qPCR.jpg\"\n width=\"200\" /></a\n ></span>\n </td>\n </tr>\n <tr>\n <td valign=\"top\">\n <p>\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><strong>SNAP-ChIP Data:</strong> Histone H3K36me3 antibody\n (5 µg) was tested in a native ChIP experiment using\n chromatin from K-562 cells (5 µg) with the SNAP-ChIP\n K-MetStat Panel (EpiCypher Catalog No. 19-1001) spiked-in\n prior to micrococcal nuclease digestion. Specificity (left\n y-axis) was determined by qPCR for the DNA barcodes\n corresponding to modified nucleosomes in the SNAP-ChIP panel\n (x-axis). Black bar represents antibody efficiency (right\n y-axis; log scale) and indicates percentage of the target\n immunoprecipitated relative to input. Error bars represent\n mean ± SEM in replicate ChIP experiments. <br />\n <strong>(Click image to enlarge) </strong></span\n >\n </p>\n\n <p></p>\n </td>\n </tr>\n <!--END IMAGE AND CAPTION SECTION-->\n <tr valign=\"top\">\n <td valign=\"top\">\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><a\n href=\"https://www.epicypher.com/content/images/products/antibodies/13-0031_seq_tracks.jpg\"\n target=\"_new\"\n ><img\n alt=\"13-0031 Representative Sequencing Tracks\"\n height=\"100\"\n src=\"https://cdn11.bigcommerce.com/s-y9o92/content/images/products/antibodies/13-0031_seq_tracks.jpg\"\n width=\"350\" /></a\n ></span>\n </td>\n </tr>\n <tr>\n <td valign=\"top\">\n <p>\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><strong\n >H3K36me3 SNAP-ChIP-seq and CUT&RUN representative tracks: </strong\n >A gene browser shot generated using the Integrative\n Genomics Viewer (IGV, Broad Institute) shows a\n representative locus for EpiCypher H3K36me3 ChIP-seq (blue\n tracks, 3 μg antibody) and CUT&RUN (green track, 1:100\n antibody dilution). For comparison ENCODE H3K36me3 ChIP-seq\n using a different antibody is shown (bottom orange track,\n GEO accession number GSM621387). Similar results in peak\n structure and location were observed throughout the genome\n for EpiCypher H3K36me3 antibody in ChIP-seq and CUT&RUN.\n <i\n >Methods: Native ChIP-seq was performed as described (Shah\n et al., Mol Cell 2018). CUT&RUN was performed using\n EpiCypher CUTANA pAG-MNase for ChIC/CUT&RUN (EpiCypher\n Catalog No. 15-1016) as described\n (EpiCypher.com/cutana-protocol). Library preparation was\n performed with 10 ng DNA using the NEBNext<sup>®</sup>\n Ultra<sup>TM</sup> II DNA Library Prep Kit for\n Illumina<sup>®</sup>. ChIP libraries were sequenced on an\n Illumina NextSeq 550 (2x150bp paired end). The total\n number of reads was 33.6 million for ChIP-seq and 3.2\n million for CUT&RUN.</i\n >\n <br />\n <strong>(Click image to enlarge) </strong></span\n >\n </p>\n\n <p></p>\n </td>\n </tr>\n <tr valign=\"top\">\n <td valign=\"top\">\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><a\n href=\"https://www.epicypher.com/content/images/products/antibodies/13-0031_GWA.jpg\"\n target=\"_new\"\n ><img\n alt=\"13-0031 Genome Wide Analysis\"\n height=\"100\"\n src=\"https://cdn11.bigcommerce.com/s-y9o92/content/images/products/antibodies/13-0031_GWA.jpg\"\n width=\"127\" /></a\n ></span>\n </td>\n </tr>\n <tr>\n <td valign=\"top\">\n <p>\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><strong>ChIP-seq and CUT&RUN genome wide analysis </strong>\n EpiCypher H3K36me3 antibody was tested in native ChIP-seq\n <strong>(A)</strong> and CUT&RUN <strong>(B)</strong> using\n the methods described above. Genome-wide analysis of\n H3K36me3 enrichment (signal intensity) flanking annotated\n genes (gene start to gene end; +/- 3kb) is graphed as a\n cumulative histogram plot (top) and shown in a heatmap\n (bottom). Individual gene loci in each row of the heatmap\n are colored by signal intensity and sorted by strongest to\n lowest enrichment (top to bottom). EpiCypher H3K36me3\n antibody displays a characteristic enrichment pattern in\n gene bodies.<br />\n <strong>(Click image to enlarge) </strong></span\n >\n </p>\n\n <p></p>\n </td>\n </tr>\n <tr valign=\"top\">\n <td valign=\"top\">\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><a\n href=\"https://www.epicypher.com/content/images/products/antibodies/13-0031_correlation.jpg\"\n target=\"_new\"\n ><img\n alt=\"13-0031 Correlation Analysis\"\n height=\"100\"\n src=\"https://cdn11.bigcommerce.com/s-y9o92/content/images/products/antibodies/13-0031_correlation.jpg\"\n width=\"127\" /></a\n ></span>\n </td>\n </tr>\n <tr>\n <td valign=\"top\">\n <p>\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><strong>ChIP-seq vs. CUT&RUN correlation analysis </strong>\n Genome-wide correlation analysis was performed to compare\n EpiCypher H3K36me3 antibody enrichment in ChIP-seq and\n CUT&RUN. The number of reads per 5 kb binned region across\n the genome is plotted for CUT&RUN (x-axis) vs. ChIP-seq\n (y-axis) (EaSeq). ChIP-seq and CUT&RUN data generated using\n this antibody are highly correlated (Pearson correlation r =\n 0.786).<br />\n <strong>(Click image to enlarge) </strong></span\n >\n </p>\n\n <p></p>\n </td>\n </tr>\n <tr valign=\"top\">\n <td valign=\"top\">\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><a\n href=\"https://www.epicypher.com/content/images/products/antibodies/13-0031_wb.jpg\"\n target=\"_new\"\n ><img\n alt=\"13-0031 Western Blot\"\n height=\"100\"\n src=\"https://cdn11.bigcommerce.com/s-y9o92/content/images/products/antibodies/13-0031_wb.jpg\"\n width=\"127\" /></a\n ></span>\n </td>\n </tr>\n <tr>\n <td valign=\"top\">\n <p>\n <span style=\"font-family: arial, helvetica, sans-serif\"\n ><strong>Western blot analysis </strong> Recombinant histone\n H3.3 (Lane 1) and acid extracts of HeLa cells (Lane 2) were\n blotted onto PVDF and probed with 2 μg/mL Histone H3K36me3\n antibody.<br />\n <strong>(Click image to enlarge) </strong></span\n >\n </p>\n\n <p></p>\n </td>\n </tr>\n </tbody>\n </table>\n </td>\n </tr>\n </tbody>\n</table>\n<style>\n .button-group {\n display: none;\n }\n</style>","tags":[],"detail_messages":"","availability":"","page_title":"Histone H3K36me3 Antibody, SNAP-ChIP® Certified, CUTANA™ CUT&RUN Compatible (13-0031)","mpn":"","upc":null,"options":[],"related_products":[{"id":411,"sku":null,"name":"SNAP-ChIP® K-MetStat Panel","url":"https://www.epicypher.com/products/nucleosomes/snap-chip-k-metstat-panel","availability":"","rating":null,"brand":{"name":null},"category":["Nucleosomes","Nucleosomes/SNAP-ChIP<sup>®</sup> Spike-ins"],"summary":"\n\t\n\t\t\n\t\t\t\n\t\t\t\n\t\t\t\t\n\t\t\t\t\t\n\t\t\t\t\t\t\n\t\t\t\t\t\t\n\n\t\t\t\t\t\t\n\t\t\t\t\t\tSNAP-ChIP K-MetStat Panel Description: A panel ","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/411/282/SNAP_Chip__53961.1569012494.png?c=2","alt":"SNAP-ChIP® K-MetStat Panel"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/411/282/SNAP_Chip__53961.1569012494.png?c=2","alt":"SNAP-ChIP® K-MetStat Panel"}],"date_added":"18th Sep 2017","pre_order":false,"show_cart_action":true,"has_options":true,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":null,"num_reviews":null,"weight":{"formatted":"0.01 LBS","value":0.01},"demo":false,"price":{"without_tax":{"currency":"USD","formatted":"$365.00","value":365},"price_range":{"min":{"without_tax":{"currency":"USD","formatted":"$365.00","value":365},"tax_label":"Sales Tax"},"max":{"without_tax":{"currency":"USD","formatted":"$2,795.00","value":2795},"tax_label":"Sales Tax"}},"tax_label":"Sales Tax"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=411"},{"id":555,"sku":null,"name":"SNAP-ChIP® OncoStat Panel","url":"https://www.epicypher.com/products/nucleosomes/snap-chip-oncostat-panel","availability":"","rating":null,"brand":{"name":null},"category":["Nucleosomes","Nucleosomes/SNAP-ChIP<sup>®</sup> Spike-ins"],"summary":" \n\n ","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/555/399/OncoStat__15594.1569012525.png?c=2","alt":"SNAP-ChIP® OncoStat Panel"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/555/399/OncoStat__15594.1569012525.png?c=2","alt":"SNAP-ChIP® OncoStat Panel"}],"date_added":"31st Jul 2018","pre_order":false,"show_cart_action":true,"has_options":true,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":null,"num_reviews":null,"weight":{"formatted":"0.01 LBS","value":0.01},"demo":false,"price":{"without_tax":{"currency":"USD","formatted":"$365.00","value":365},"price_range":{"min":{"without_tax":{"currency":"USD","formatted":"$365.00","value":365},"tax_label":"Sales Tax"},"max":{"without_tax":{"currency":"USD","formatted":"$2,795.00","value":2795},"tax_label":"Sales Tax"}},"tax_label":"Sales Tax"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=555"},{"id":626,"sku":"13-0028","name":"Histone H3K4me3 Antibody, SNAP-ChIP Certified *DISCONTINUED*","url":"https://www.epicypher.com/products/antibodies/snap-chip-certified-antibodies/histone-h3k4me3-antibody-snap-chip-certified","availability":"","rating":null,"brand":{"name":null},"category":[],"summary":"\n \n \n \n ","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/626/578/Screen_Shot_2018-09-25_at_2.00.27_PM__52796.1538135192.png?c=2","alt":"Histone H3K4me3 Antibody, SNAP-ChIP Certified *DISCONTINUED*"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/626/578/Screen_Shot_2018-09-25_at_2.00.27_PM__52796.1538135192.png?c=2","alt":"Histone H3K4me3 Antibody, SNAP-ChIP Certified *DISCONTINUED*"}],"date_added":"24th Sep 2018","pre_order":false,"show_cart_action":false,"has_options":false,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":[{"id":452,"name":"Pack Size","value":"100 μg"}],"num_reviews":null,"weight":{"formatted":"0.00 LBS","value":0},"demo":false,"add_to_wishlist_url":"/wishlist.php?action=add&product_id=626"},{"id":625,"sku":"13-0027","name":"Histone H3K4me2 Antibody, SNAP-Certified™ for CUT&RUN","url":"https://www.epicypher.com/products/antibodies/cutana-cut-run-antibodies/cut-run-antibodies-histone-ptms/histone-h3k4me2-antibody-snap-certified-for-cut-run","availability":"","rating":null,"brand":{"name":null},"category":["Antibodies/CUTANA™ CUT&RUN Antibodies","Antibodies/CUTANA™ CUT&RUN Antibodies/CUTANA™ CUT&RUN Antibodies to Histone PTMs","Epigenetics Kits and Reagents/CUTANA™ ChIC / CUT&RUN Assays"],"summary":"\n \n \n Type: Monoclonal\n \n \n Target Size: 15 kDa\n \n \n Format: Affi","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/625/577/Screen_Shot_2018-09-25_at_2.00.27_PM__04625.1538135192.png?c=2","alt":"Histone H3K4me2 Antibody, SNAP-Certified™ for CUT&RUN"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/625/577/Screen_Shot_2018-09-25_at_2.00.27_PM__04625.1538135192.png?c=2","alt":"Histone H3K4me2 Antibody, SNAP-Certified™ for CUT&RUN"}],"date_added":"24th Sep 2018","pre_order":false,"show_cart_action":true,"has_options":false,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":[{"id":451,"name":"Pack Size","value":"100 μg"}],"num_reviews":null,"weight":{"formatted":"0.00 LBS","value":0},"demo":false,"add_to_cart_url":"https://www.epicypher.com/cart.php?action=add&product_id=625","price":{"without_tax":{"currency":"USD","formatted":"$525.00","value":525},"tax_label":"Sales Tax"},"add_to_wishlist_url":"/wishlist.php?action=add&product_id=625"},{"id":628,"sku":"13-0030","name":"Histone H3K27me3 Antibody, SNAP-ChIP® Certified, CUTANA™ CUT&RUN Compatible *DISCONTINUED*","url":"https://www.epicypher.com/products/antibodies/snap-chip-certified-antibodies/histone-h3k27me3-antibody-snap-chip-certified-cutana-cut-run-compatible-discontinued","availability":"","rating":null,"brand":{"name":null},"category":[],"summary":"\n \n ","image":{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/628/940/histone-h3k27me3-antibody-snap-chip-certified-cutana-cutandrun-compatible-discontinued__17318.1645734531.jpg?c=2","alt":"Histone H3K27me3 Antibody, SNAP-ChIP Certified, CUTANA CUTandRUN Compatible - DISCONTINUED"},"images":[{"data":"https://cdn11.bigcommerce.com/s-y9o92/images/stencil/{:size}/products/628/940/histone-h3k27me3-antibody-snap-chip-certified-cutana-cutandrun-compatible-discontinued__17318.1645734531.jpg?c=2","alt":"Histone H3K27me3 Antibody, SNAP-ChIP Certified, CUTANA CUTandRUN Compatible - DISCONTINUED"}],"date_added":"24th Sep 2018","pre_order":false,"show_cart_action":false,"has_options":false,"stock_level":null,"low_stock_level":null,"qty_in_cart":0,"custom_fields":[{"id":470,"name":"Pack Size","value":"100 μg"}],"num_reviews":null,"weight":{"formatted":"0.00 LBS","value":0},"demo":false,"add_to_wishlist_url":"/wishlist.php?action=add&product_id=628"}],"shipping_messages":[],"rating":0,"title":"Histone H3K36me3 Antibody, SNAP-ChIP® Certified, CUTANA™ CUT&RUN Compatible *DISCONTINUED*","gift_wrapping_available":false,"min_purchase_quantity":0} Pack Size: 100 μg
Current stock:
0